Global dissection of alternative splicing uncovers transcriptional diversity in tissues and associates with the flavonoid pathway in tea plant (Camellia sinensis)
- PMID: 30400863
- PMCID: PMC6219262
- DOI: 10.1186/s12870-018-1497-9
Global dissection of alternative splicing uncovers transcriptional diversity in tissues and associates with the flavonoid pathway in tea plant (Camellia sinensis)
Abstract
Background: Alternative splicing (AS) regulates mRNA at the post-transcriptional level to change gene function in organisms. However, little is known about the AS and its roles in tea plant (Camellia sinensis), widely cultivated for making a popular beverage tea.
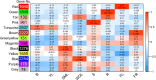
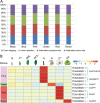
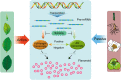
Results: In our study, the AS landscape and dynamics were characterized in eight tissues (bud, young leaf, summer mature leaf, winter old leaf, stem, root, flower, fruit) of tea plant by Illumina RNA-Seq and confirmed by Iso-Seq. The most abundant AS (~ 20%) was intron retention and involved in RNA processes. The some alternative splicings were found to be tissue specific in stem and root etc. Thirteen co-expressed modules of AS transcripts were identified, which revealed a similar pattern between the bud and young leaves as well as a distinct pattern between seasons. AS events of structural genes including anthocyanidin reductase and MYB transcription factors were involved in biosynthesis of flavonoid, especially in vegetative tissues. The AS isoforms rather than the full-length ones were the major transcripts involved in flavonoid synthesis pathway, and is positively correlated with the catechins content conferring the tea taste. We propose that the AS is an important functional mechanism in regulating flavonoid metabolites.
Conclusion: Our study provides the insight into the AS events underlying tea plant's uniquely different developmental process and highlights the important contribution and efficacy of alternative splicing regulatory function to biosynthesis of flavonoids.
Keywords: Alternative splicing; Camellia sinensis; Flavonoid; Tissue-specificity.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not Applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis).BMC Genomics. 2015 Jul 29;16(1):560. doi: 10.1186/s12864-015-1773-0. BMC Genomics. 2015. PMID: 26220550 Free PMC article.
-
Comprehensive identification of the full-length transcripts and alternative splicing related to the secondary metabolism pathways in the tea plant (Camellia sinensis).Sci Rep. 2019 Feb 25;9(1):2709. doi: 10.1038/s41598-019-39286-z. Sci Rep. 2019. PMID: 30804390 Free PMC article.
-
Comprehensive co-expression analysis provides novel insights into temporal variation of flavonoids in fresh leaves of the tea plant (Camellia sinensis).Plant Sci. 2020 Jan;290:110306. doi: 10.1016/j.plantsci.2019.110306. Epub 2019 Oct 15. Plant Sci. 2020. PMID: 31779914
-
Flavonoid metabolites in tea plant (Camellia sinensis) stress response: Insights from bibliometric analysis.Plant Physiol Biochem. 2023 Sep;202:107934. doi: 10.1016/j.plaphy.2023.107934. Epub 2023 Aug 3. Plant Physiol Biochem. 2023. PMID: 37572493 Review.
-
Anthocyanin metabolism and its differential regulation in purple tea (Camellia sinensis).Plant Physiol Biochem. 2023 Aug;201:107875. doi: 10.1016/j.plaphy.2023.107875. Epub 2023 Jul 9. Plant Physiol Biochem. 2023. PMID: 37451003 Review.
Cited by
-
Genome-wide identification of the PYL gene family of tea plants (Camellia sinensis) revealed its expression profiles under different stress and tissues.BMC Genomics. 2023 Jun 28;24(1):362. doi: 10.1186/s12864-023-09464-5. BMC Genomics. 2023. PMID: 37380940 Free PMC article.
-
Intron-retained radish (Raphanus sativus L.) RsMYB1 transcripts found in colored-taproot lines enhance anthocyanin accumulation in transgenic Arabidopsis plants.Plant Cell Rep. 2021 Sep;40(9):1735-1749. doi: 10.1007/s00299-021-02735-z. Epub 2021 Jul 25. Plant Cell Rep. 2021. PMID: 34308490
-
Brassinosteroid Priming Improves Peanut Drought Tolerance via Eliminating Inhibition on Genes in Photosynthesis and Hormone Signaling.Genes (Basel). 2020 Aug 11;11(8):919. doi: 10.3390/genes11080919. Genes (Basel). 2020. PMID: 32796553 Free PMC article.
-
Alternative Splicing and Its Roles in Plant Metabolism.Int J Mol Sci. 2022 Jul 1;23(13):7355. doi: 10.3390/ijms23137355. Int J Mol Sci. 2022. PMID: 35806361 Free PMC article. Review.
-
Identification and distribution of a single nucleotide polymorphism responsible for the catechin content in tea plants.Hortic Res. 2020 Mar 1;7:24. doi: 10.1038/s41438-020-0247-y. eCollection 2020. Hortic Res. 2020. PMID: 32140233 Free PMC article.
References
-
- Shi CY, Yang H, Wei CL, Yu O, Zhang ZZ, Jiang CJ, Sun J, Li YY, Chen Q, Xia T, Wan XC. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 2011;12:131. doi: 10.1186/1471-2164-12-131. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 31171608/National Natural Science Foundation of China
- 15czs08032/Special Innovative Province Construction in Anhui Province
- 2016080503B024/Special Project for Central Guiding Science and Technology Innovatio of Region in Anhui Province
- IRT1101/Programme for Changjiang Scholars and Innovative Research Team in University
LinkOut - more resources
Full Text Sources
Research Materials

