Extracellular vesicles derived from natural killer cells use multiple cytotoxic proteins and killing mechanisms to _target cancer cells
- PMID: 30891164
- PMCID: PMC6419691
- DOI: 10.1080/20013078.2019.1588538
Extracellular vesicles derived from natural killer cells use multiple cytotoxic proteins and killing mechanisms to _target cancer cells
Abstract
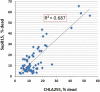
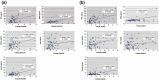
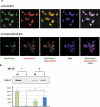
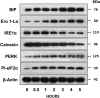
Extracellular vesicles (EVs) are secreted membrane vesicles, which play complex physiological and pathological functions in intercellular communication. Recently, we isolated natural killer (NK) cell-derived EVs (NK-EVs) from ex vivo expansion of NK cell cultures. The isolated NK-EVs contained cytotoxic proteins and several activated caspases, and they induced apoptosis in _target cells. In this report, the protein levels of cytotoxic proteins from NK-EV isolates were analysed by ELISA. The mean values of perforin (PFN, 550 ng/mL), granzyme A (GzmA, 185 ng/mL), granzyme B (GzmB, 23.4 ng/mL), granulysin (GNLY, 56 ng/mL), and FasL (2.5 ng/mL) were obtained from >60 isolations using dot plots. The correlation between cytotoxicity and cytotoxic protein levels was examined by linear regression. PFN, GzmA, GzmB, GNLY all had a positive, moderate correlation with cytotoxicity, suggesting that there is not a single cytotoxic protein dominantly involved in killing and that all of these proteins may contribute to cytotoxicity. To further explore the possible killing mechanisms, cells were treated with NK-EVs, proteins extracted and lysates assessed by Western blotting. The levels of Gzm A substrates, SET and HMG2, were diminished in _targeted cells, indicating that GzmA may induce a caspase-independent death pathway. Also, cytochrome C was released from mitochondria, a central hallmark of caspase-dependent death pathways. In addition, several ER-associated proteins were altered, suggesting that NK-EVs may induce ER stress resulting in cell death. Our results indicate that multiple killing mechanisms are activated by NK-derived EVs, including caspase-independent and -dependent cell death pathways, which can mediate cytotoxicity against cancer cells. Abbreviations: NK: natural killer cells; aNK: activated NK cells; EV: extracellular vesicles; ER: endoplasmic reticulum; ALL: acute lymphoblastic leukaemia; FBS: foetal bovine serum. GzmA: granzyme A; GzmB: granzyme B; GNLY: granulysin; PFN: perforin.
Keywords: Scale-up isolation; cancer treatment; caspases; cytotoxicity; extracellular vesicles; natural killer cells.
Figures



Similar articles
-
Large-scale isolation and cytotoxicity of extracellular vesicles derived from activated human natural killer cells.J Extracell Vesicles. 2017 Feb 28;6(1):1294368. doi: 10.1080/20013078.2017.1294368. eCollection 2017. J Extracell Vesicles. 2017. PMID: 28326171 Free PMC article.
-
Evaluation of resazurin phenoxazine dye as a highly sensitive cell viability potency assay for natural killer cell-derived extracellular vesicle-based cancer biotherapeutics.J Extracell Biol. 2024 Jul 17;3(7):e166. doi: 10.1002/jex2.166. eCollection 2024 Jul. J Extracell Biol. 2024. PMID: 39022723 Free PMC article.
-
Combined Role of Interleukin-15 Stimulated Natural Killer Cell-Derived Extracellular Vesicles and Carboplatin in Osimertinib-Resistant H1975 Lung Cancer Cells with EGFR Mutations.Pharmaceutics. 2024 Jan 8;16(1):83. doi: 10.3390/pharmaceutics16010083. Pharmaceutics. 2024. PMID: 38258094 Free PMC article.
-
Professional killers: The role of extracellular vesicles in the reciprocal interactions between natural killer, CD8+ cytotoxic T-cells and tumour cells.J Extracell Vesicles. 2021 Apr;10(6):e12075. doi: 10.1002/jev2.12075. Epub 2021 Apr 1. J Extracell Vesicles. 2021. PMID: 33815694 Free PMC article. Review.
-
New mechanism of organophosphorus pesticide-induced immunotoxicity.J Nippon Med Sch. 2007 Apr;74(2):92-105. doi: 10.1272/jnms.74.92. J Nippon Med Sch. 2007. PMID: 17507786 Review.
Cited by
-
Extracellular vesicles in anti-tumor drug resistance: Mechanisms and therapeutic prospects.J Pharm Anal. 2024 Jul;14(7):100920. doi: 10.1016/j.jpha.2023.12.010. Epub 2023 Dec 16. J Pharm Anal. 2024. PMID: 39104866 Free PMC article. Review.
-
Extracellular Vesicles and Immune System Function: Exploring Novel Approaches to Colorectal Cancer Immunotherapy.Biomedicines. 2024 Jul 3;12(7):1473. doi: 10.3390/biomedicines12071473. Biomedicines. 2024. PMID: 39062046 Free PMC article. Review.
-
Extracellular Vesicles and Their Role in the Spatial and Temporal Expansion of Tumor-Immune Interactions.Int J Mol Sci. 2021 Mar 25;22(7):3374. doi: 10.3390/ijms22073374. Int J Mol Sci. 2021. PMID: 33806053 Free PMC article. Review.
-
Multiplex plasma protein assays as a diagnostic tool for lung cancer.Cancer Sci. 2024 Oct;115(10):3439-3454. doi: 10.1111/cas.16300. Epub 2024 Jul 30. Cancer Sci. 2024. PMID: 39080998 Free PMC article.
-
Extracellular vesicles and cancer stem cells: a deadly duo in tumor progression.Oncol Rev. 2024 Jul 18;18:1411736. doi: 10.3389/or.2024.1411736. eCollection 2024. Oncol Rev. 2024. PMID: 39091989 Free PMC article. Review.
References
-
- Bobrie A, Colombo M, Raposo G, et al. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12(12):1659–13. Epub 2011/ 06/08 PubMed PMID: 21645191. - PubMed
-
- Théry C, Zitvogel L, Amigorena S.. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. .PubMed PMID: 12154376 - PubMed
-
- Camussi G, Deregibus MC, Bruno S, et al. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78(9):838–848. Epub 2010/08/13 PubMed PMID: 20703216. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

