An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5
- PMID: 30990354
- PMCID: PMC6984602
- DOI: 10.1080/15548627.2019.1606637
An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5
Abstract
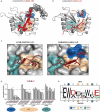
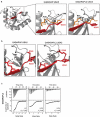
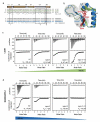
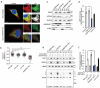
Short linear motifs, known as LC3-interacting regions (LIRs), interact with mactoautophagy/autophagy modifiers (Atg8/LC3/GABARAP proteins) via a conserved universal mechanism. Typically, this includes the occupancy of 2 hydrophobic pockets on the surface of Atg8-family proteins by 2 specific aromatic and hydrophobic residues within the LIR motifs. Here, we describe an alternative mechanism of Atg8-family protein interaction with the non-canonical UBA5 LIR, an E1-like enzyme of the ufmylation pathway that preferentially interacts with GABARAP but not LC3 proteins. By solving the structures of both GABARAP and GABARAPL2 in complex with the UBA5 LIR, we show that in addition to the binding to the 2 canonical hydrophobic pockets (HP1 and HP2), a conserved tryptophan residue N-terminal of the LIR core sequence binds into a novel hydrophobic pocket on the surface of GABARAP proteins, which we term HP0. This mode of action is unique for UBA5 and accompanied by large rearrangements of key residues including the side chains of the gate-keeping K46 and the adjacent K/R47 in GABARAP proteins. Swapping mutations in LC3B and GABARAPL2 revealed that K/R47 is the key residue in the specific binding of GABARAP proteins to UBA5, with synergetic contributions of the composition and dynamics of the loop L3. Finally, we elucidate the physiological relevance of the interaction and show that GABARAP proteins regulate the localization and function of UBA5 on the endoplasmic reticulum membrane in a lipidation-independent manner.Abbreviations: ATG: AuTophaGy-related; EGFP: enhanced green fluorescent protein; GABARAP: GABA-type A receptor-associated protein; ITC: isothermal titration calorimetry; KO: knockout; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; NMR: nuclear magnetic resonance; RMSD: root-mean-square deviation of atomic positions; TKO: triple knockout; UBA5: ubiquitin like modifier activating enzyme 5.
Keywords: Autophagy; GABARAP; LC3; LIR; UFM1; complex structure; endoplasmic reticulum; peptide arrays; ufmylation.
Figures
Similar articles
-
Structural and Functional Analysis of a Novel Interaction Motif within UFM1-activating Enzyme 5 (UBA5) Required for Binding to Ubiquitin-like Proteins and Ufmylation.J Biol Chem. 2016 Apr 22;291(17):9025-41. doi: 10.1074/jbc.M116.715474. Epub 2016 Feb 29. J Biol Chem. 2016. PMID: 26929408 Free PMC article.
-
Human LC3 and GABARAP subfamily members achieve functional specificity via specific structural modulations.Autophagy. 2020 Feb;16(2):239-255. doi: 10.1080/15548627.2019.1606636. Epub 2019 Apr 28. Autophagy. 2020. PMID: 30982432 Free PMC article.
-
Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.Autophagy. 2019 Aug;15(8):1333-1355. doi: 10.1080/15548627.2019.1581009. Epub 2019 Mar 4. Autophagy. 2019. PMID: 30767700 Free PMC article.
-
Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors.J Mol Biol. 2020 Jan 3;432(1):80-103. doi: 10.1016/j.jmb.2019.07.016. Epub 2019 Jul 13. J Mol Biol. 2020. PMID: 31310766 Review.
-
Mechanisms of Selective Autophagy.Annu Rev Cell Dev Biol. 2021 Oct 6;37:143-169. doi: 10.1146/annurev-cellbio-120219-035530. Epub 2021 Jun 21. Annu Rev Cell Dev Biol. 2021. PMID: 34152791 Review.
Cited by
-
Shuffled ATG8 interacting motifs form an ancestral bridge between UFMylation and autophagy.EMBO J. 2023 May 15;42(10):e112053. doi: 10.15252/embj.2022112053. Epub 2023 Feb 10. EMBO J. 2023. PMID: 36762703 Free PMC article.
-
The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.Nat Commun. 2022 Dec 21;13(1):7857. doi: 10.1038/s41467-022-35501-0. Nat Commun. 2022. PMID: 36543799 Free PMC article.
-
Highly Specialized Ubiquitin-Like Modifications: Shedding Light into the UFM1 Enigma.Biomolecules. 2021 Feb 10;11(2):255. doi: 10.3390/biom11020255. Biomolecules. 2021. PMID: 33578803 Free PMC article. Review.
-
Dysregulation of ribosome-associated quality control elicits cognitive disorders via overaccumulation of TTC3.Proc Natl Acad Sci U S A. 2023 Mar 21;120(12):e2211522120. doi: 10.1073/pnas.2211522120. Epub 2023 Mar 14. Proc Natl Acad Sci U S A. 2023. PMID: 36917672 Free PMC article.
-
Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5.Commun Biol. 2021 Mar 5;4(1):291. doi: 10.1038/s42003-021-01830-x. Commun Biol. 2021. PMID: 33674710 Free PMC article.
References
-
- Mizushima N, Komatsu M.. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. - PubMed
-
- Rogov V, Dötsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. - PubMed
-
- Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem. 2017;86:193–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
