Superovulation alters global DNA methylation in early mouse embryo development
- PMID: 31060426
- PMCID: PMC6615540
- DOI: 10.1080/15592294.2019.1615353
Superovulation alters global DNA methylation in early mouse embryo development
Abstract
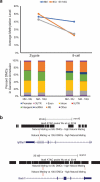
Assisted reproductive technologies are known to alter the developmental environment of gametes and early embryos during the most dynamic period of establishing the epigenome. This may result in the introduction of errors during active DNA methylation reprogramming. Controlled ovarian hyperstimulation, or superovulation, is a ubiquitously used intervention which has been demonstrated to alter the methylation of certain imprinted genes. The objective of this study was to investigate whether ovarian hyperstimulation results in genome-wide DNA methylation changes in mouse early embryos. Ovarian hyperstimulation was induced by treating mice with either low doses (5 IU) or high doses (10 IU) of PMSG and hCG. Natural mating (NM) control mice received no treatment. Zygotes and 8-cell embryos were collected from each group and DNA methylomes were generated by whole-genome bisulfite sequencing. In the NM group, mean CpG methylation levels slightly decreased from zygote to 8-cell stage, whereas a large decrease in mean CpG methylation level was observed in both superovulated groups. A separate analysis of the mean CpG methylation levels within each developmental stage confirmed that significant genome-wide erasure of CpG methylation from the zygote to 8-cell stage only occurred in the superovulation groups. Our results suggest that superovulation alters the genome-wide DNA methylation erasure process in mouse early pre-implantation embryos. It is not clear whether these changes are transient or persistent. Further studies are ongoing to investigate the impact of ovarian hyperstimulation on DNA methylation re-establishment in later stages of embryo development.
Keywords: DNA methylation; assisted reproductive technologies; embryo development; epigenetics; ovarian hyperstimulation.
Figures

Similar articles
-
Genome-wide assessment of DNA methylation alterations induced by superovulation, sexual immaturity and in vitro follicle growth in mouse blastocysts.Clin Epigenetics. 2023 Jan 16;15(1):9. doi: 10.1186/s13148-023-01421-z. Clin Epigenetics. 2023. PMID: 36647174 Free PMC article.
-
Perturbations in imprinted methylation from assisted reproductive technologies but not advanced maternal age in mouse preimplantation embryos.Clin Epigenetics. 2019 Nov 26;11(1):162. doi: 10.1186/s13148-019-0751-9. Clin Epigenetics. 2019. PMID: 31767035 Free PMC article.
-
Single-cell DNA methylation sequencing reveals epigenetic alterations in mouse oocytes superovulated with different dosages of gonadotropins.Clin Epigenetics. 2020 Jun 1;12(1):75. doi: 10.1186/s13148-020-00866-w. Clin Epigenetics. 2020. PMID: 32487258 Free PMC article.
-
The effects of superovulation and reproductive aging on the epigenome of the oocyte and embryo.Mol Reprod Dev. 2018 Feb;85(2):90-105. doi: 10.1002/mrd.22951. Epub 2018 Jan 22. Mol Reprod Dev. 2018. PMID: 29280527 Review.
-
DNA methylation reprogramming during oogenesis and interference by reproductive technologies: Studies in mouse and bovine models.Reprod Fertil Dev. 2015 Jun;27(5):739-54. doi: 10.1071/RD14333. Reprod Fertil Dev. 2015. PMID: 25976160 Review.
Cited by
-
DNA methylation status of imprinted H19 and KvDMR1 genes in human placentas after conception using assisted reproductive technology.Ann Transl Med. 2020 Jul;8(14):854. doi: 10.21037/atm-20-3364. Ann Transl Med. 2020. PMID: 32793698 Free PMC article.
-
Evaluating histone modification analysis of individual preimplantation embryos.BMC Genomics. 2024 Jan 18;25(1):75. doi: 10.1186/s12864-024-09984-8. BMC Genomics. 2024. PMID: 38238676 Free PMC article.
-
Temporary Increased LDL-C in Offspring with Extreme Elevation of Maternal Preconception Estradiol: A Retrospective Cohort Study.Clin Epidemiol. 2022 Apr 7;14:453-462. doi: 10.2147/CLEP.S358999. eCollection 2022. Clin Epidemiol. 2022. PMID: 35418784 Free PMC article.
-
Placental Abnormalities are Associated With Specific Windows of Embryo Culture in a Mouse Model.Front Cell Dev Biol. 2022 Apr 25;10:884088. doi: 10.3389/fcell.2022.884088. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35547813 Free PMC article.
-
Effects of gonadotropin administration on clinical outcomes in clomiphene citrate-based minimal stimulation cycle IVF.Reprod Med Biol. 2019 Dec 12;19(2):128-134. doi: 10.1002/rmb2.12310. eCollection 2020 Apr. Reprod Med Biol. 2019. PMID: 32273817 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
