Trypanosoma brucei ribonuclease H2A is an essential R-loop processing enzyme whose loss causes DNA damage during transcription initiation and antigenic variation
- PMID: 31350892
- PMCID: PMC6753483
- DOI: 10.1093/nar/gkz644
Trypanosoma brucei ribonuclease H2A is an essential R-loop processing enzyme whose loss causes DNA damage during transcription initiation and antigenic variation
Abstract
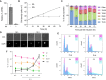
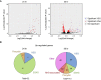
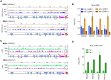
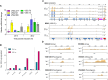
Ribonucleotides represent a threat to DNA genome stability and transmission. Two types of Ribonuclease H (RNase H) excise ribonucleotides when they form part of the DNA strand, or hydrolyse RNA when it base-pairs with DNA in structures termed R-loops. Loss of either RNase H is lethal in mammals, whereas yeast survives the absence of both enzymes. RNase H1 loss is tolerated by the parasite Trypanosoma brucei but no work has examined the function of RNase H2. Here we show that loss of T. brucei RNase H2 (TbRH2A) leads to growth and cell cycle arrest that is concomitant with accumulation of nuclear damage at sites of RNA polymerase (Pol) II transcription initiation, revealing a novel and critical role for RNase H2. Differential gene expression analysis reveals limited overall changes in RNA levels for RNA Pol II genes after TbRH2A loss, but increased perturbation of nucleotide metabolic genes. Finally, we show that TbRH2A loss causes R-loop and DNA damage accumulation in telomeric RNA Pol I transcription sites, also leading to altered gene expression. Thus, we demonstrate separation of function between two nuclear T. brucei RNase H enzymes during RNA Pol II transcription, but overlap in function during RNA Pol I-mediated gene expression during host immune evasion.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
A DOT1B/Ribonuclease H2 Protein Complex Is Involved in R-Loop Processing, Genomic Integrity, and Antigenic Variation in Trypanosoma brucei.mBio. 2021 Dec 21;12(6):e0135221. doi: 10.1128/mBio.01352-21. Epub 2021 Nov 9. mBio. 2021. PMID: 34749530 Free PMC article.
-
Ribonuclease H1-_targeted R-loops in surface antigen gene expression sites can direct trypanosome immune evasion.PLoS Genet. 2018 Dec 13;14(12):e1007729. doi: 10.1371/journal.pgen.1007729. eCollection 2018 Dec. PLoS Genet. 2018. PMID: 30543624 Free PMC article.
-
Genome-wide mapping reveals conserved and diverged R-loop activities in the unusual genetic landscape of the African trypanosome genome.Nucleic Acids Res. 2018 Dec 14;46(22):11789-11805. doi: 10.1093/nar/gky928. Nucleic Acids Res. 2018. PMID: 30304482 Free PMC article.
-
RNases H1 and H2: guardians of the stability of the nuclear genome when supply of dNTPs is limiting for DNA synthesis.Curr Genet. 2020 Dec;66(6):1073-1084. doi: 10.1007/s00294-020-01086-8. Epub 2020 Sep 4. Curr Genet. 2020. PMID: 32886170 Free PMC article. Review.
-
The contribution of chromosomal translocations to antigenic variation in Trypanosoma brucei.Philos Trans R Soc Lond B Biol Sci. 1984 Nov 13;307(1131):13-26. doi: 10.1098/rstb.1984.0105. Philos Trans R Soc Lond B Biol Sci. 1984. PMID: 6151678 Review.
Cited by
-
An SR protein is essential for activating DNA repair in malaria parasites.J Cell Sci. 2021 Aug 15;134(16):jcs258572. doi: 10.1242/jcs.258572. Epub 2021 Aug 17. J Cell Sci. 2021. PMID: 34291805 Free PMC article.
-
Unpicking the Roles of DNA Damage Protein Kinases in Trypanosomatids.Front Cell Dev Biol. 2021 Aug 6;9:636615. doi: 10.3389/fcell.2021.636615. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34422791 Free PMC article. Review.
-
RAD51-mediated R-loop formation acts to repair transcription-associated DNA breaks driving antigenic variation in Trypanosoma brucei.Proc Natl Acad Sci U S A. 2023 Nov 28;120(48):e2309306120. doi: 10.1073/pnas.2309306120. Epub 2023 Nov 21. Proc Natl Acad Sci U S A. 2023. PMID: 37988471 Free PMC article.
-
A DOT1B/Ribonuclease H2 Protein Complex Is Involved in R-Loop Processing, Genomic Integrity, and Antigenic Variation in Trypanosoma brucei.mBio. 2021 Dec 21;12(6):e0135221. doi: 10.1128/mBio.01352-21. Epub 2021 Nov 9. mBio. 2021. PMID: 34749530 Free PMC article.
-
Escaping the immune system by DNA repair and recombination in African trypanosomes.Open Biol. 2019 Nov 29;9(11):190182. doi: 10.1098/rsob.190182. Epub 2019 Nov 13. Open Biol. 2019. PMID: 31718509 Free PMC article. Review.
References
-
- Traut T.W. Physiological concentrations of purines and pyrimidines. Mol. Cell Biochem. 1994; 140:1–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/M028909/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 104111/WT_/Wellcome Trust/United Kingdom
- BB/K006495/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/N016165/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

