Inhibition of the ULK1 protein complex suppresses Staphylococcus-induced autophagy and cell death
- PMID: 31387948
- PMCID: PMC6768650
- DOI: 10.1074/jbc.RA119.008923
Inhibition of the ULK1 protein complex suppresses Staphylococcus-induced autophagy and cell death
Abstract
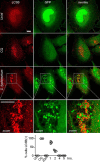
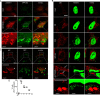
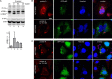
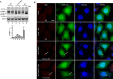
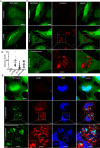
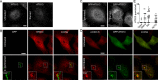
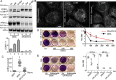
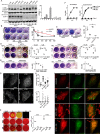
Autophagy plays multiple roles in host cells challenged with extracellular pathogens. Here, we aimed to explore whether autophagy inhibition could prevent bacterial infections. We first confirmed widely distinct patterns of autophagy responses in host cells infected with Staphylococcus aureus, as compared with Salmonella Only infection with Staphylococcus produced strong accumulation of lipidated autophagy-related protein LC3B (LC3B-II). Infection with virulent Staphylococcus strains induced formation of p62-positive aggregates, suggestive of accumulated ubiquitinated _targets. During Salmonella infection, bacteria remain enclosed by lysosomal-associated membrane protein 2 (LAMP2)-positive lysosomes, whereas virulent Staphylococcus apparently exited from enlarged lysosomes and invaded the cytoplasm. Surprisingly, Staphylococcus appeared to escape from the lysosome without generation of membrane-damage signals as detected by galectin-3 recruitment. In contrast, Salmonella infection produced high levels of lysosomal damage, consistent with a downstream antibacterial xenophagy response. Finally, we studied the Unc-51-like autophagy-activating kinase 1 (ULK1) regulatory complex, including the essential subunit autophagy-related protein 13 (ATG13). Infection of cells with either Staphylococcus or Salmonella led to recruitment of ATG13 to sites of cytosolic bacterial cells to promote autophagosome formation. Of note, genetic _targeting of ATG13 suppressed autophagy and the ability of Staphylococcus to infect and kill host cells. Two different ULK1 inhibitors also prevented Staphylococcus intracellular replication and host cell death. Interestingly, inhibition of the ULK1 pathway had the opposite effect on Salmonella, sensitizing cells to the infection. Our results suggest that ULK1 inhibitors may offer a potential strategy to impede cellular infection by S. aureus.
Keywords: Salmonella enterica; Staphylococcus aureus (S. aureus); Unc-51-like autophagy-activating kinase 1 (ULK1); autophagy; autophagy-related protein 13 (ATG13); bacterial pathogenesis; bacterial virulence; infection; intracellular pathogen; xenophagy.
© 2019 Radhi et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
Poliovirus induces autophagic signaling independent of the ULK1 complex.Autophagy. 2018;14(7):1201-1213. doi: 10.1080/15548627.2018.1458805. Epub 2018 Jul 20. Autophagy. 2018. PMID: 29929428 Free PMC article.
-
The unc-51 like autophagy activating kinase 1-autophagy related 13 complex has distinct functions in tunicamycin-treated cells.Biochem Biophys Res Commun. 2020 Apr 9;524(3):744-749. doi: 10.1016/j.bbrc.2020.01.160. Epub 2020 Feb 5. Biochem Biophys Res Commun. 2020. PMID: 32035621
-
AMPK Inhibits ULK1-Dependent Autophagosome Formation and Lysosomal Acidification via Distinct Mechanisms.Mol Cell Biol. 2018 Apr 30;38(10):e00023-18. doi: 10.1128/MCB.00023-18. Print 2018 May 15. Mol Cell Biol. 2018. PMID: 29507183 Free PMC article.
-
The mammalian ULK1 complex and autophagy initiation.Essays Biochem. 2017 Dec 12;61(6):585-596. doi: 10.1042/EBC20170021. Print 2017 Dec 12. Essays Biochem. 2017. PMID: 29233870 Free PMC article. Review.
-
Regulation of Autophagy through TORC1 and mTORC1.Biomolecules. 2017 Jul 7;7(3):52. doi: 10.3390/biom7030052. Biomolecules. 2017. PMID: 28686223 Free PMC article. Review.
Cited by
-
A Review of ULK1-Mediated Autophagy in Drug Resistance of Cancer.Cancers (Basel). 2020 Feb 4;12(2):352. doi: 10.3390/cancers12020352. Cancers (Basel). 2020. PMID: 32033142 Free PMC article. Review.
-
Effects of the linoleic acid/docosahexaenoic acid ratio and concentration inducing autophagy in Raw264.7 cells against Staphylococcus aureus.J Clin Biochem Nutr. 2020 Sep;67(2):146-152. doi: 10.3164/jcbn.19-95. Epub 2020 Apr 17. J Clin Biochem Nutr. 2020. PMID: 33041511 Free PMC article.
-
Autophagy in Crohn's Disease: Converging on Dysfunctional Innate Immunity.Cells. 2023 Jul 4;12(13):1779. doi: 10.3390/cells12131779. Cells. 2023. PMID: 37443813 Free PMC article. Review.
-
Making the Most of the Host; _targeting the Autophagy Pathway Facilitates Staphylococcus aureus Intracellular Survival in Neutrophils.Front Immunol. 2021 Jun 16;12:667387. doi: 10.3389/fimmu.2021.667387. eCollection 2021. Front Immunol. 2021. PMID: 34220813 Free PMC article. Review.
-
Competitive Cell Death Interactions in Pulmonary Infection: Host Modulation Versus Pathogen Manipulation.Front Immunol. 2020 May 19;11:814. doi: 10.3389/fimmu.2020.00814. eCollection 2020. Front Immunol. 2020. PMID: 32508813 Free PMC article. Review.
References
-
- Zhao Z., Fux B., Goodwin M., Dunay I. R., Strong D., Miller B. C., Cadwell K., Delgado M. A., Ponpuak M., Green K. G., Schmidt R. E., Mizushima N., Deretic V., Sibley L. D., and Virgin H. W. (2008) Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 10.1016/j.chom.2008.10.003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

