Discovery and Characterization of Novel RNA Viruses in Aquatic North American Wild Birds
- PMID: 31438486
- PMCID: PMC6784231
- DOI: 10.3390/v11090768
Discovery and Characterization of Novel RNA Viruses in Aquatic North American Wild Birds
Abstract
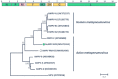
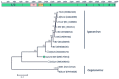
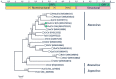
Wild birds are recognized viral reservoirs but our understanding about avian viral diversity is limited. We describe here three novel RNA viruses that we identified in oropharyngeal/cloacal swabs collected from wild birds. The complete genome of a novel gull metapneumovirus (GuMPV B29) was determined. Phylogenetic analyses indicated that this virus could represent a novel avian metapneumovirus (AMPV) sub-group, intermediate between AMPV-C and the subgroup of the other AMPVs. This virus was detected in an American herring (1/24, 4.2%) and great black-backed (4/26, 15.4%) gulls. A novel gull coronavirus (GuCoV B29) was detected in great black-backed (3/26, 11.5%) and American herring (2/24, 8.3%) gulls. Phylogenetic analyses of GuCoV B29 suggested that this virus could represent a novel species within the genus Gammacoronavirus, close to other recently identified potential novel avian coronaviral species. One GuMPV-GuCoV co-infection was detected. A novel duck calicivirus (DuCV-2 B6) was identified in mallards (2/5, 40%) and American black ducks (7/26, 26.9%). This virus, of which we identified two different types, was fully sequenced and was genetically closest to other caliciviruses identified in Anatidae, but more distant to other caliciviruses from birds in the genus Anas. These discoveries increase our knowledge about avian virus diversity and host distributions.
Keywords: avian viruses; calicivirus; coronavirus; metapneumovirus; novel viruses; viral epidemiology; virus discovery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Avian metapneumovirus subtype C in Wild Waterfowl in Ontario, Canada.Transbound Emerg Dis. 2018 Aug;65(4):1098-1102. doi: 10.1111/tbed.12832. Epub 2018 Feb 18. Transbound Emerg Dis. 2018. PMID: 29457370
-
Diversity of Coronaviruses in Wild Representatives of the Aves Class in Poland.Viruses. 2021 Jul 29;13(8):1497. doi: 10.3390/v13081497. Viruses. 2021. PMID: 34452362 Free PMC article.
-
Variation in viral shedding patterns between different wild bird species infected experimentally with low-pathogenicity avian influenza viruses that originated from wild birds.Avian Pathol. 2011 Apr;40(2):119-24. doi: 10.1080/03079457.2010.540002. Avian Pathol. 2011. PMID: 21500030
-
Epidemiology of low pathogenic avian influenza viruses in wild birds.Rev Sci Tech. 2009 Apr;28(1):49-58. doi: 10.20506/rst.28.1.1863. Rev Sci Tech. 2009. PMID: 19618618 Review.
-
[An overview of surveillance of avian influenza viruses in wild birds].Bing Du Xue Bao. 2014 May;30(3):310-7. Bing Du Xue Bao. 2014. PMID: 25118388 Review. Chinese.
Cited by
-
Disentangling the role of wild birds in avian metapneumovirus (aMPV) epidemiology: A systematic review and meta-analysis.Transbound Emerg Dis. 2022 Nov;69(6):3285-3299. doi: 10.1111/tbed.14680. Epub 2022 Aug 22. Transbound Emerg Dis. 2022. PMID: 35960706 Free PMC article.
-
Investigating the Diversity and Host Range of Novel Parvoviruses from North American Ducks Using Epidemiology, Phylogenetics, Genome Structure, and Codon Usage Analysis.Viruses. 2021 Jan 28;13(2):193. doi: 10.3390/v13020193. Viruses. 2021. PMID: 33525386 Free PMC article.
-
Rapid loss of maternal immunity and increase in environmentally mediated antibody generation in urban gulls.Sci Rep. 2024 Feb 22;14(1):4357. doi: 10.1038/s41598-024-54796-1. Sci Rep. 2024. PMID: 38388645 Free PMC article.
-
Coronaviruses in the Sea.Front Microbiol. 2020 Jul 24;11:1795. doi: 10.3389/fmicb.2020.01795. eCollection 2020. Front Microbiol. 2020. PMID: 32793180 Free PMC article. Review.
-
Introduction of Avian metapneumovirus subtype A to the United States: molecular insights and implications.Front Microbiol. 2024 Jul 5;15:1428248. doi: 10.3389/fmicb.2024.1428248. eCollection 2024. Front Microbiol. 2024. PMID: 39035438 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

