Genome-wide Profiling of Histone Lysine Butyrylation Reveals its Role in the Positive Regulation of Gene Transcription in Rice
- PMID: 31776817
- PMCID: PMC6881504
- DOI: 10.1186/s12284-019-0342-6
Genome-wide Profiling of Histone Lysine Butyrylation Reveals its Role in the Positive Regulation of Gene Transcription in Rice
Abstract
Background: Histone modifications play important roles in growth and development of rice (Oryza sativa L.). Lysine butyrylation (Kbu) with a four-carbon chain is a newly-discovered histone acylation modification in rice.
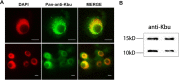
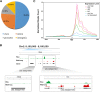
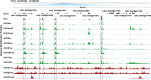
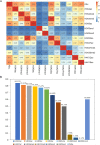
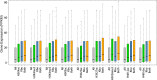
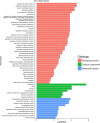
Main body: In this study, we performed chromatin immunoprecipitation sequencing (ChIP-seq) analyses, the result showed that major enrichment of histone Kbu located in genebody regions of rice genome, especially in exons. The enrichment level of Kbu histone modification is positively correlated with gene expression. Furthermore, we compared Kbu with DNase-seq and other histone modifications in rice. We found that 60.06% Kub enriched region co-located with DHSs in intergenic regions. The similar profiles were detected among Kbu and several acetylation modifications such as H3K4ac, H3K9ac, and H3K23ac, indicating that Kbu modification is an active signal of transcription. Genes with both histone Kbu and one other acetylation also had significantly increased expression compared with genes with only one acetylation. Gene Ontology (GO) enrichment analysis revealed that these genes with histone Kbu can regulate multiple metabolic process in different rice varieties.
Conclusion: Our study showed that the lysine butyrylation modificaiton may promote gene expression as histone acetylation and will provide resources for futher studies on histone Kbu and other epigenetic modifications in plants.
Keywords: ChIP-seq; Histone modification; Lysine butyrylation (Kbu); Oryza sativa; Transcriptional regulation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Functions and mechanisms of protein lysine butyrylation (Kbu): Therapeutic implications in human diseases.Genes Dis. 2022 Nov 29;10(6):2479-2490. doi: 10.1016/j.gendis.2022.10.025. eCollection 2023 Nov. Genes Dis. 2022. PMID: 37554202 Free PMC article. Review.
-
Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence.Genome Biol. 2018 Sep 25;19(1):144. doi: 10.1186/s13059-018-1533-y. Genome Biol. 2018. PMID: 30253806 Free PMC article.
-
Global Involvement of Lysine Crotonylation in Protein Modification and Transcription Regulation in Rice.Mol Cell Proteomics. 2018 Oct;17(10):1922-1936. doi: 10.1074/mcp.RA118.000640. Epub 2018 Jul 18. Mol Cell Proteomics. 2018. PMID: 30021883 Free PMC article.
-
Genome-wide analysis of histone modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. Japonica.Mol Plant. 2013 Sep;6(5):1463-72. doi: 10.1093/mp/sst018. Epub 2013 Jan 25. Mol Plant. 2013. PMID: 23355544 Free PMC article.
-
Epigenetic landscape of amphetamine and methamphetamine addiction in rodents.Epigenetics. 2015;10(7):574-80. doi: 10.1080/15592294.2015.1055441. Epigenetics. 2015. PMID: 26023847 Free PMC article. Review.
Cited by
-
Functions and mechanisms of protein lysine butyrylation (Kbu): Therapeutic implications in human diseases.Genes Dis. 2022 Nov 29;10(6):2479-2490. doi: 10.1016/j.gendis.2022.10.025. eCollection 2023 Nov. Genes Dis. 2022. PMID: 37554202 Free PMC article. Review.
-
Crossing epigenetic frontiers: the intersection of novel histone modifications and diseases.Signal Transduct _target Ther. 2024 Sep 16;9(1):232. doi: 10.1038/s41392-024-01918-w. Signal Transduct _target Ther. 2024. PMID: 39278916 Free PMC article. Review.
-
Insight into the Role of Epigenetic Processes in Abiotic and Biotic Stress Response in Wheat and Barley.Int J Mol Sci. 2020 Feb 21;21(4):1480. doi: 10.3390/ijms21041480. Int J Mol Sci. 2020. PMID: 32098241 Free PMC article. Review.
-
First comprehensive analysis of lysine succinylation in paper mulberry (Broussonetia papyrifera).BMC Genomics. 2021 Apr 10;22(1):255. doi: 10.1186/s12864-021-07567-5. BMC Genomics. 2021. PMID: 33838656 Free PMC article.
-
Epigenetic Regulation in Response to CO2 Fluctuation in Marine Microalga Nannochloropsis oceanica.Microb Ecol. 2023 Nov 28;87(1):4. doi: 10.1007/s00248-023-02322-7. Microb Ecol. 2023. PMID: 38015286
References
Grants and funding
LinkOut - more resources
Full Text Sources

