Evolution of imprinting via lineage-specific insertion of retroviral promoters
- PMID: 31831741
- PMCID: PMC6908575
- DOI: 10.1038/s41467-019-13662-9
Evolution of imprinting via lineage-specific insertion of retroviral promoters
Abstract
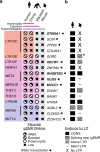
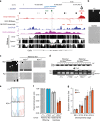
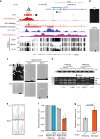
Imprinted genes are expressed from a single parental allele, with the other allele often silenced by DNA methylation (DNAme) established in the germline. While species-specific imprinted orthologues have been documented, the molecular mechanisms underlying the evolutionary switch from biallelic to imprinted expression are unknown. During mouse oogenesis, gametic differentially methylated regions (gDMRs) acquire DNAme in a transcription-guided manner. Here we show that oocyte transcription initiating in lineage-specific endogenous retroviruses (ERVs) is likely responsible for DNAme establishment at 4/6 mouse-specific and 17/110 human-specific imprinted gDMRs. The latter are divided into Catarrhini- or Hominoidea-specific gDMRs embedded within transcripts initiating in ERVs specific to these primate lineages. Strikingly, imprinting of the maternally methylated genes Impact and Slc38a4 was lost in the offspring of female mice harboring deletions of the relevant murine-specific ERVs upstream of these genes. Our work reveals an evolutionary mechanism whereby maternally silenced genes arise from biallelically expressed progenitors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues.Genome Biol. 2019 Oct 29;20(1):225. doi: 10.1186/s13059-019-1833-x. Genome Biol. 2019. PMID: 31665063 Free PMC article.
-
Maternal regulation of chromosomal imprinting in animals.Chromosoma. 2019 Jun;128(2):69-80. doi: 10.1007/s00412-018-00690-5. Epub 2019 Feb 5. Chromosoma. 2019. PMID: 30719566 Free PMC article. Review.
-
Angelman syndrome imprinting center encodes a transcriptional promoter.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):6871-5. doi: 10.1073/pnas.1411261111. Epub 2014 Nov 5. Proc Natl Acad Sci U S A. 2015. PMID: 25378697 Free PMC article.
-
Germline and somatic imprinting in the nonhuman primate highlights species differences in oocyte methylation.Genome Res. 2015 May;25(5):611-23. doi: 10.1101/gr.183301.114. Epub 2015 Apr 10. Genome Res. 2015. PMID: 25862382 Free PMC article.
-
Cross-species clues of an epigenetic imprinting regulatory code for the IGF2R gene.Cytogenet Genome Res. 2006;113(1-4):202-8. doi: 10.1159/000090833. Cytogenet Genome Res. 2006. PMID: 16575181 Review.
Cited by
-
Host Gene Regulation by Transposable Elements: The New, the Old and the Ugly.Viruses. 2020 Sep 26;12(10):1089. doi: 10.3390/v12101089. Viruses. 2020. PMID: 32993145 Free PMC article. Review.
-
Noncanonical imprinting sustains embryonic development and restrains placental overgrowth.Genes Dev. 2022 Apr 1;36(7-8):483-494. doi: 10.1101/gad.349390.122. Epub 2022 Apr 28. Genes Dev. 2022. PMID: 35483741 Free PMC article.
-
Distinctive aspects of the placental epigenome and theories as to how they arise.Cell Mol Life Sci. 2022 Oct 26;79(11):569. doi: 10.1007/s00018-022-04568-9. Cell Mol Life Sci. 2022. PMID: 36287261 Free PMC article. Review.
-
Maternal H3K27me3-dependent autosomal and X chromosome imprinting.Nat Rev Genet. 2020 Sep;21(9):555-571. doi: 10.1038/s41576-020-0245-9. Epub 2020 Jun 8. Nat Rev Genet. 2020. PMID: 32514155 Free PMC article. Review.
-
The enigma of DNA methylation in the mammalian oocyte.F1000Res. 2020 Feb 25;9:F1000 Faculty Rev-146. doi: 10.12688/f1000research.21513.1. eCollection 2020. F1000Res. 2020. PMID: 32148772 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

