BAP18 is involved in upregulation of CCND1/2 transcription to promote cell growth in oral squamous cell carcinoma
- PMID: 32113162
- PMCID: PMC7047197
- DOI: 10.1016/j.ebiom.2020.102685
BAP18 is involved in upregulation of CCND1/2 transcription to promote cell growth in oral squamous cell carcinoma
Abstract
Background: As a reader of histone H3K4me3, BPTF associated protein of 18 kDa (BAP18) is involved in modulation of androgen receptor action in prostate cancer. However, the function of BAP18 on oral squamous cell carcinoma (OSCC) and its molecular mechanism remains to be elusive.
Methods: OSCC-derived cell lines carrying silenced BAP18 were established by Lentiviral infection. Quantitative PCR (qPCR), western blot, and ChIP assay were performed to detect gene transcription regulation and the possible mechanism. Colony formation, cell growth curve and xenograft tumor experiments were performed to examine cell growth and proliferation.
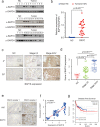
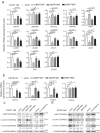
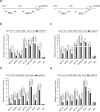
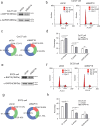
Findings: Our study demonstrated that BAP18 was highly expressed in OSCC samples compared with that in benign. BAP18 depletion obviously influenced the expression of a series of genes, including cell cycle-related genes. We thus provided the evidence to demonstrate that BAP18 depletion significantly decreases CCND1 and CCND2 (CCND1/2) transcription. In addition, BAP18 is recruited to the promoter regions of CCND1/2, thereby facilitating the recruitment of the core subunits of MLL1 complex to the same regions, to increase histone H3K4me3 levels. Furthermore, BAP18 depletion delayed G1-S phase transition and inhibited cell growth in OSCC-derived cell lines.
Interpretation: This study suggests that BAP18 is involved in modulation of CCND1/2 transcription and promotes OSCC progression. BAP18 could be a potential _target for OSCC treatment and diagnosis. FUND: This work was funded by National Natural Science Foundation of China (31871286, 81872015, 31701102, 81702800, 81902889), Foundation for Special Professor of Liaoning Province, and Supported project for young technological innovation-talents in Shenyang (No. RC170541).
Keywords: BAP18; CCND1/2; Histone methylation; Oral squamous cell carcinoma; Transcription regulation.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interests.
Figures

Similar articles
-
BAP18 induces growth of non-small-cell lung carcinoma through upregulating transcriptional level of CCND1/2.Eur Rev Med Pharmacol Sci. 2022 May;26(9):3074-3082. doi: 10.26355/eurrev_202205_28724. Eur Rev Med Pharmacol Sci. 2022. PMID: 35587057
-
Upregulation of miR-519d-3p Inhibits Viability, Proliferation, and G1/S Cell Cycle Transition of Oral Squamous Cell Carcinoma Cells Through _targeting CCND1.Cancer Biother Radiopharm. 2024 Mar;39(2):153-163. doi: 10.1089/cbr.2020.3984. Epub 2020 Oct 14. Cancer Biother Radiopharm. 2024. PMID: 33052706
-
Dlx5 promotes cancer progression through regulation of CCND1 in oral squamous cell carcinoma (OSCC).Biochem Cell Biol. 2021 Aug;99(4):424-434. doi: 10.1139/bcb-2020-0523. Epub 2021 Jul 20. Biochem Cell Biol. 2021. PMID: 34283652
-
Knocking-down of CREPT prohibits the progression of oral squamous cell carcinoma and suppresses cyclin D1 and c-Myc expression.PLoS One. 2017 Apr 3;12(4):e0174309. doi: 10.1371/journal.pone.0174309. eCollection 2017. PLoS One. 2017. PMID: 28369091 Free PMC article.
-
An update on the implications of cyclin D1 in oral carcinogenesis.Oral Dis. 2017 Oct;23(7):897-912. doi: 10.1111/odi.12620. Epub 2017 Mar 31. Oral Dis. 2017. PMID: 27973759 Review.
Cited by
-
Long non-coding RNA TFAP2A-AS1 plays an important role in oral squamous cell carcinoma: research includes bioinformatics analysis and experiments.BMC Oral Health. 2022 May 6;22(1):160. doi: 10.1186/s12903-022-02203-4. BMC Oral Health. 2022. PMID: 35524329 Free PMC article.
-
Chromatin complexes subunit BAP18 promotes triple-negative breast cancer progression through transcriptional activation of oncogene S100A9.Cell Death Dis. 2022 Apr 28;13(4):408. doi: 10.1038/s41419-022-04785-x. Cell Death Dis. 2022. PMID: 35484101 Free PMC article.
-
The Ash2l SDI Domain Is Required to Maintain the Stability and Binding of DPY30.Cells. 2022 Apr 25;11(9):1450. doi: 10.3390/cells11091450. Cells. 2022. PMID: 35563756 Free PMC article.
-
Circ_0000745 strengthens the expression of CCND1 by functioning as miR-488 sponge and interacting with HuR binding protein to facilitate the development of oral squamous cell carcinoma.Cancer Cell Int. 2021 May 21;21(1):271. doi: 10.1186/s12935-021-01884-1. Cancer Cell Int. 2021. PMID: 34020639 Free PMC article.
-
BAP18 facilitates CTCF-mediated chromatin accessible to regulate enhancer activity in breast cancer.Cell Death Differ. 2023 May;30(5):1260-1278. doi: 10.1038/s41418-023-01135-y. Epub 2023 Feb 24. Cell Death Differ. 2023. PMID: 36828916 Free PMC article.
References
-
- Chuang S.C., Scelo G., Tonita J.M., Tamaro S., Jonasson J.G., Kliewer E.V. Risk of second primary cancer among patients with head and neck cancers: a pooled analysis of 13 cancer registries. Int J Cancer. 2008;123(10):2390–2396. - PubMed
-
- Sant M., Aareleid T., Berrino F., Bielska Lasota M., Carli P.M., Faivre J. EUROCARE-3: survival of cancer patients diagnosed 1990-94–results and commentary. Ann Oncol. 2003;14(Suppl 5):v61–118. - PubMed
-
- Patel S.C., Carpenter W.R., Tyree S., Couch M.E., Weissler M., Hackman T. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J Clin Oncol. 2011;29(11):1488–1494. - PubMed
-
- Ng J.H., Iyer N.G., Tan M.H., Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck. 2017;39(2):297–304. - PubMed
-
- van Dijk B.A., Brands M.T., Geurts S.M., Merkx M.A., Roodenburg J.L. Trends in oral cavity cancer incidence, mortality, survival and treatment in the Netherlands. Int J Cancer. 2016;139(3):574–583. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

