Transient receptor potential ion channel TRPM2 promotes AML proliferation and survival through modulation of mitochondrial function, ROS, and autophagy
- PMID: 32312983
- PMCID: PMC7170900
- DOI: 10.1038/s41419-020-2454-8
Transient receptor potential ion channel TRPM2 promotes AML proliferation and survival through modulation of mitochondrial function, ROS, and autophagy
Abstract
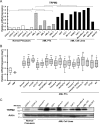
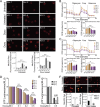
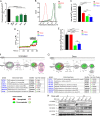
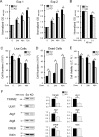
Transient receptor potential melastatin 2 (TRPM2) ion channel has an essential function in maintaining cell survival following oxidant injury. Here, we show that TRPM2 is highly expressed in acute myeloid leukemia (AML). The role of TRPM2 in AML was studied following depletion with CRISPR/Cas9 technology in U937 cells. In in vitro experiments and in xenografts, depletion of TRPM2 in AML inhibited leukemia proliferation, and doxorubicin sensitivity was increased. Mitochondrial function including oxygen consumption rate and ATP production was reduced, impairing cellular bioenergetics. Mitochondrial membrane potential and mitochondrial calcium uptake were significantly decreased in depleted cells. Mitochondrial reactive oxygen species (ROS) were significantly increased, and Nrf2 was decreased, reducing the antioxidant response. In TRPM2-depleted cells, ULK1, Atg7, and Atg5 protein levels were decreased, leading to autophagy inhibition. Consistently, ATF4 and CREB, two master transcription factors for autophagosome biogenesis, were reduced in TRPM2-depleted cells. In addition, Atg13 and FIP200, which are known to stabilize ULK1 protein, were decreased. Reconstitution with TRPM2 fully restored proliferation, viability, and autophagy; ATF4 and CREB fully restored proliferation and viability but only partially restored autophagy. TRPM2 expression reduced the elevated ROS found in depleted cells. These data show that TRPM2 has an important role in AML proliferation and survival through regulation of key transcription factors and _target genes involved in mitochondrial function, bioenergetics, the antioxidant response, and autophagy. _targeting TRPM2 may represent a novel therapeutic approach to inhibit myeloid leukemia growth and enhance susceptibility to chemotherapeutic agents through multiple pathways.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Depletion of the Human Ion Channel TRPM2 in Neuroblastoma Demonstrates Its Key Role in Cell Survival through Modulation of Mitochondrial Reactive Oxygen Species and Bioenergetics.J Biol Chem. 2016 Nov 18;291(47):24449-24464. doi: 10.1074/jbc.M116.747147. Epub 2016 Sep 30. J Biol Chem. 2016. PMID: 27694440 Free PMC article.
-
The human ion channel TRPM2 modulates neuroblastoma cell survival and mitochondrial function through Pyk2, CREB, and MCU activation.Am J Physiol Cell Physiol. 2018 Oct 1;315(4):C571-C586. doi: 10.1152/ajpcell.00098.2018. Epub 2018 Jul 18. Am J Physiol Cell Physiol. 2018. PMID: 30020827 Free PMC article.
-
TRPM2 channel-mediated regulation of autophagy maintains mitochondrial function and promotes gastric cancer cell survival via the JNK-signaling pathway.J Biol Chem. 2018 Mar 9;293(10):3637-3650. doi: 10.1074/jbc.M117.817635. Epub 2018 Jan 17. J Biol Chem. 2018. PMID: 29343514 Free PMC article.
-
TRPM2 in Cancer.Cell Calcium. 2019 Jun;80:8-17. doi: 10.1016/j.ceca.2019.03.002. Epub 2019 Mar 6. Cell Calcium. 2019. PMID: 30925291 Free PMC article. Review.
-
Transient Receptor Potential-Melastatin Channel Family Member 2: Friend or Foe.Trans Am Clin Climatol Assoc. 2017;128:308-329. Trans Am Clin Climatol Assoc. 2017. PMID: 28790515 Free PMC article. Review.
Cited by
-
CD38-NADase is a new major contributor to Duchenne muscular dystrophic phenotype.EMBO Mol Med. 2022 May 9;14(5):e12860. doi: 10.15252/emmm.202012860. Epub 2022 Mar 17. EMBO Mol Med. 2022. PMID: 35298089 Free PMC article.
-
Transient Receptor Potential (TRP) Channels in Haematological Malignancies: An Update.Biomolecules. 2021 May 20;11(5):765. doi: 10.3390/biom11050765. Biomolecules. 2021. PMID: 34065398 Free PMC article. Review.
-
_targeting Transient Receptor Potential Melastatin-2 (TRPM2) Enhances Therapeutic Efficacy of Third Generation EGFR Inhibitors against EGFR Mutant Lung Cancer.Adv Sci (Weinh). 2024 Sep;11(35):e2310126. doi: 10.1002/advs.202310126. Epub 2024 Jul 23. Adv Sci (Weinh). 2024. PMID: 39044361 Free PMC article.
-
Electroacupuncture pretreatment alleviates myocardial injury through regulating mitochondrial function.Eur J Med Res. 2020 Aug 1;25(1):29. doi: 10.1186/s40001-020-00431-4. Eur J Med Res. 2020. PMID: 32738910 Free PMC article.
-
Put in a "Ca2+ll" to Acute Myeloid Leukemia.Cells. 2022 Feb 4;11(3):543. doi: 10.3390/cells11030543. Cells. 2022. PMID: 35159351 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

