Metabolism and the Epigenome: A Dynamic Relationship
- PMID: 32387193
- PMCID: PMC8477637
- DOI: 10.1016/j.tibs.2020.04.002
Metabolism and the Epigenome: A Dynamic Relationship
Abstract
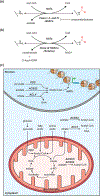
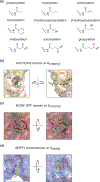
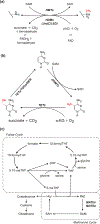
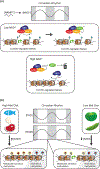
Many chromatin-modifying enzymes require metabolic cofactors to support their catalytic activities, providing a direct path for fluctuations in metabolite availability to regulate the epigenome. Over the past decade, our knowledge of this link has grown significantly. What began with studies showing that cofactor availability drives global abundances of chromatin modifications has transitioned to discoveries highlighting metabolic enzymes as loci-specific regulators of gene expression. Here, we cover our current understanding of mechanisms that facilitate the dynamic and complex relationship between metabolism and the epigenome, focusing on the roles of essential metabolic and chromatin associated enzymes. We discuss physiological conditions where availability of these epimetabolites is dynamically altered, highlighting known links to the epigenome and proposing other plausible connections.
Keywords: acetylation; acylation; chromatin; circadian cycles; dietary challenges; methylation.
Copyright © 2020. Published by Elsevier Ltd.
Figures

Similar articles
-
Genome-wide investigation of the dynamic changes of epigenome modifications after global DNA methylation editing.Nucleic Acids Res. 2021 Jan 11;49(1):158-176. doi: 10.1093/nar/gkaa1169. Nucleic Acids Res. 2021. PMID: 33300025 Free PMC article.
-
Metabolic regulation of the plant epigenome.Plant J. 2023 Jun;114(5):1001-1013. doi: 10.1111/tpj.16122. Epub 2023 Feb 10. Plant J. 2023. PMID: 36705504 Review.
-
Histone acylations and chromatin dynamics: concepts, challenges, and links to metabolism.EMBO Rep. 2021 Jul 5;22(7):e52774. doi: 10.15252/embr.202152774. Epub 2021 Jun 23. EMBO Rep. 2021. PMID: 34159701 Free PMC article. Review.
-
Regulation of the epigenome through RNA modifications.Chromosoma. 2023 Sep;132(3):231-246. doi: 10.1007/s00412-023-00794-7. Epub 2023 May 4. Chromosoma. 2023. PMID: 37138119 Free PMC article. Review.
-
Metabolism and epigenetics: a link cancer cells exploit.Curr Opin Biotechnol. 2015 Aug;34:23-9. doi: 10.1016/j.copbio.2014.11.012. Epub 2014 Nov 29. Curr Opin Biotechnol. 2015. PMID: 25461508 Free PMC article. Review.
Cited by
-
Unraveling the epigenetic code: human kidney DNA methylation and chromatin dynamics in renal disease development.Nat Commun. 2024 Jan 29;15(1):873. doi: 10.1038/s41467-024-45295-y. Nat Commun. 2024. PMID: 38287030 Free PMC article.
-
The molecular athlete: exercise physiology from mechanisms to medals.Physiol Rev. 2023 Jul 1;103(3):1693-1787. doi: 10.1152/physrev.00017.2022. Epub 2023 Jan 5. Physiol Rev. 2023. PMID: 36603158 Free PMC article. Review.
-
Metabolism in the Midwest: research from the Midwest Aging Consortium at the 49th Annual Meeting of the American Aging Association.Geroscience. 2022 Feb;44(1):39-52. doi: 10.1007/s11357-021-00479-y. Epub 2021 Oct 29. Geroscience. 2022. PMID: 34714522 Free PMC article. No abstract available.
-
Metabolic Control of Germline Formation and Differentiation in Mammals.Sex Dev. 2022;16(5-6):388-403. doi: 10.1159/000520662. Epub 2022 Jan 27. Sex Dev. 2022. PMID: 35086109 Free PMC article. Review.
-
Mitochondria signaling to the epigenome: A novel role for an old organelle.Free Radic Biol Med. 2021 Jul;170:59-69. doi: 10.1016/j.freeradbiomed.2020.11.016. Epub 2020 Dec 1. Free Radic Biol Med. 2021. PMID: 33271282 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

