Integration of high-throughput reporter assays identify a critical enhancer of the Ikzf1 gene
- PMID: 32453736
- PMCID: PMC7250416
- DOI: 10.1371/journal.pone.0233191
Integration of high-throughput reporter assays identify a critical enhancer of the Ikzf1 gene
Erratum in
-
Correction: Integration of high-throughput reporter assays identify a critical enhancer of the Ikzf1 gene.PLoS One. 2020 Jun 25;15(6):e0235578. doi: 10.1371/journal.pone.0235578. eCollection 2020. PLoS One. 2020. PMID: 32584906 Free PMC article.
Abstract
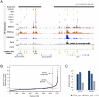
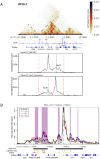
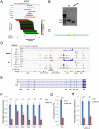
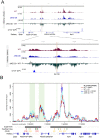
The Ikzf1 locus encodes the lymphoid specific transcription factor Ikaros, which plays an essential role in both T and B cell differentiation, while deregulation or mutation of IKZF1/Ikzf1 is involved in leukemia. Tissue-specific and cell identity genes are usually associated with clusters of enhancers, also called super-enhancers, which are believed to ensure proper regulation of gene expression throughout cell development and differentiation. Several potential regulatory regions have been identified in close proximity of Ikzf1, however, the full extent of the regulatory landscape of the Ikzf1 locus is not yet established. In this study, we combined epigenomics and transcription factor binding along with high-throughput enhancer assay and 4C-seq to prioritize an enhancer element located 120 kb upstream of the Ikzf1 gene. We found that deletion of the E120 enhancer resulted in a significant reduction of Ikzf1 mRNA. However, the epigenetic landscape and 3D topology of the locus were only slightly affected, highlighting the complexity of the regulatory landscape regulating the Ikzf1 locus.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures
Similar articles
-
Transcriptional regulation of the Ikzf1 locus.Blood. 2013 Oct 31;122(18):3149-59. doi: 10.1182/blood-2013-01-474916. Epub 2013 Sep 3. Blood. 2013. PMID: 24002445 Free PMC article.
-
Enhancer polymorphisms at the IKZF1 susceptibility locus for acute lymphoblastic leukemia impact B-cell proliferation and differentiation in both Down syndrome and non-Down syndrome genetic backgrounds.PLoS One. 2021 Jan 7;16(1):e0244863. doi: 10.1371/journal.pone.0244863. eCollection 2021. PLoS One. 2021. PMID: 33411777 Free PMC article.
-
Odd-skipped related 1 gene expression is regulated by Runx2 and Ikzf1 transcription factors.Gene. 2008 Dec 15;426(1-2):81-90. doi: 10.1016/j.gene.2008.08.015. Epub 2008 Sep 3. Gene. 2008. PMID: 18804520
-
The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia.Haematologica. 2018 Apr;103(4):565-574. doi: 10.3324/haematol.2017.185603. Epub 2018 Mar 8. Haematologica. 2018. PMID: 29519871 Free PMC article. Review.
-
Ikaros and leukaemia.Br J Haematol. 2015 May;169(4):479-91. doi: 10.1111/bjh.13342. Epub 2015 Mar 5. Br J Haematol. 2015. PMID: 25753742 Review.
Cited by
-
Correction: Integration of high-throughput reporter assays identify a critical enhancer of the Ikzf1 gene.PLoS One. 2020 Jun 25;15(6):e0235578. doi: 10.1371/journal.pone.0235578. eCollection 2020. PLoS One. 2020. PMID: 32584906 Free PMC article.
-
From Genotype to Phenotype: How Enhancers Control Gene Expression and Cell Identity in Hematopoiesis.Hemasphere. 2023 Nov 8;7(11):e969. doi: 10.1097/HS9.0000000000000969. eCollection 2023 Nov. Hemasphere. 2023. PMID: 37953829 Free PMC article. Review.
-
Ikaros Proteins in Tumor: Current Perspectives and New Developments.Front Mol Biosci. 2021 Dec 7;8:788440. doi: 10.3389/fmolb.2021.788440. eCollection 2021. Front Mol Biosci. 2021. PMID: 34950704 Free PMC article. Review.
-
A Comprehensive Toolbox to Analyze Enhancer-Promoter Functions.Methods Mol Biol. 2021;2351:3-22. doi: 10.1007/978-1-0716-1597-3_1. Methods Mol Biol. 2021. PMID: 34382181 Review.
References
-
- Bevington SL, Cauchy P, Cockerill PN. Chromatin priming elements establish immunological memory in T cells without activating transcription: T cell memory is maintained by DNA elements which stably prime inducible genes without activating steady state transcription. BioEssays: news and reviews in molecular, cellular and developmental biology. 2017;39(2). Epub 2016/12/28. 10.1002/bies.201600184 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

