RETREG1/FAM134B mediated autophagosomal degradation of AMFR/GP78 and OPA1 -a dual organellar turnover mechanism
- PMID: 32559118
- PMCID: PMC8354597
- DOI: 10.1080/15548627.2020.1783118
RETREG1/FAM134B mediated autophagosomal degradation of AMFR/GP78 and OPA1 -a dual organellar turnover mechanism
Abstract
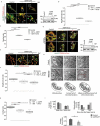
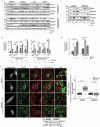
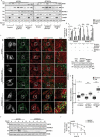
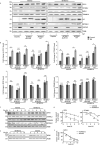
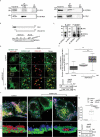
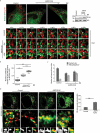
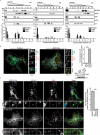
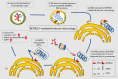
Turnover of cellular organelles, including endoplasmic reticulum (ER) and mitochondria, is orchestrated by an efficient cellular surveillance system. We have identified a mechanism for dual regulation of ER and mitochondria under stress. It is known that AMFR, an ER E3 ligase and ER-associated degradation (ERAD) regulator, degrades outer mitochondrial membrane (OMM) proteins, MFNs (mitofusins), via the proteasome and triggers mitophagy. We show that destabilized mitochondria are almost devoid of the OMM and generate "mitoplasts". This brings the inner mitochondrial membrane (IMM) in the proximity of the ER. When AMFR levels are high and the mitochondria are stressed, the reticulophagy regulatory protein RETREG1 participates in the formation of the mitophagophore by interacting with OPA1. Interestingly, OPA1 and other IMM proteins exhibit similar RETREG1-dependent autophagosomal degradation as AMFR, unlike most of the OMM proteins. The "mitoplasts" generated are degraded by reticulo-mito-phagy - simultaneously affecting dual organelle turnover.Abbreviations: AMFR/GP78: autocrine motility factor receptor; BAPTA: 1,2-bis(o-aminophenoxy)ethane-N,N,N',N'-tetraacetic acid; BFP: blue fluorescent protein; CCCP: carbonyl cyanide m-chlorophenyl hydrazone; CNBr: cyanogen bromide; ER: endoplasmic reticulum; ERAD: endoplasmic-reticulum-associated protein degradation; FL: fluorescence, GFP: green fluorescent protein; HA: hemagglutinin; HEPES: 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; IMM: inner mitochondrial membrane; LIR: LC3-interacting region; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MFN: mitofusin, MGRN1: mahogunin ring finger 1; NA: numerical aperature; OMM: outer mitochondrial membrane; OPA1: OPA1 mitochondrial dynamin like GTPase; PRNP/PrP: prion protein; RER: rough endoplasmic reticulum; RETREG1/FAM134B: reticulophagy regulator 1; RFP: red fluorescent protein; RING: really interesting new gene; ROI: region of interest; RTN: reticulon; SEM: standard error of the mean; SER: smooth endoplasmic reticulum; SIM: structured illumination microscopy; SQSTM1/p62: sequestosome 1; STED: stimulated emission depletion; STOML2: stomatin like 2; TOMM20: translocase of outer mitochondrial membrane 20; UPR: unfolded protein response.
Keywords: AMFR/GP78; OPA1; RETREG1/FAM134B; autophagy; mitoplast; reticulo-mito-phagy.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures

Similar articles
-
USP20 deubiquitinates and stabilizes the reticulophagy receptor RETREG1/FAM134B to drive reticulophagy.Autophagy. 2024 Aug;20(8):1780-1797. doi: 10.1080/15548627.2024.2347103. Epub 2024 May 12. Autophagy. 2024. PMID: 38705724 Free PMC article.
-
Organelle-specific autophagy in inflammatory diseases: a potential therapeutic _target underlying the quality control of multiple organelles.Autophagy. 2021 Feb;17(2):385-401. doi: 10.1080/15548627.2020.1725377. Epub 2020 Feb 12. Autophagy. 2021. PMID: 32048886 Free PMC article. Review.
-
Regulation of mitophagy by the Gp78 E3 ubiquitin ligase.Mol Biol Cell. 2013 Apr;24(8):1153-62. doi: 10.1091/mbc.E12-08-0607. Epub 2013 Feb 20. Mol Biol Cell. 2013. PMID: 23427266 Free PMC article.
-
Distinct mechanisms controlling rough and smooth endoplasmic reticulum contacts with mitochondria.J Cell Sci. 2015 Aug 1;128(15):2759-65. doi: 10.1242/jcs.171132. Epub 2015 Jun 11. J Cell Sci. 2015. PMID: 26065430
-
Reticulophagy and viral infection.Autophagy. 2025 Jan;21(1):3-20. doi: 10.1080/15548627.2024.2414424. Epub 2024 Oct 23. Autophagy. 2025. PMID: 39394962 Free PMC article. Review.
Cited by
-
ER-Phagy, ER Homeostasis, and ER Quality Control: Implications for Disease.Trends Biochem Sci. 2021 Aug;46(8):630-639. doi: 10.1016/j.tibs.2020.12.013. Epub 2021 Jan 25. Trends Biochem Sci. 2021. PMID: 33509650 Free PMC article. Review.
-
ISGylation of DRP1 closely balances other post-translational modifications to mediate mitochondrial fission.Cell Death Dis. 2024 Mar 2;15(3):184. doi: 10.1038/s41419-024-06543-7. Cell Death Dis. 2024. PMID: 38431611 Free PMC article.
-
Autophagy: Regulator of cell death.Cell Death Dis. 2023 Oct 4;14(10):648. doi: 10.1038/s41419-023-06154-8. Cell Death Dis. 2023. PMID: 37794028 Free PMC article. Review.
-
Molecular characterization of wild-type and HSAN2B-linked FAM134B.Mol Biol Rep. 2023 Jul;50(7):6005-6017. doi: 10.1007/s11033-023-08517-y. Epub 2023 Jun 5. Mol Biol Rep. 2023. PMID: 37273064
-
Expression analysis and functional characterization of thioredoxin domain-containing protein 11.Mol Biol Rep. 2022 Nov;49(11):10541-10556. doi: 10.1007/s11033-022-07932-x. Epub 2022 Sep 24. Mol Biol Rep. 2022. PMID: 36152228
References
-
- Mizushima N.Autophagy: process and function. Genes Dev. 2007;21:2861–2873. - PubMed
-
- Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12:1535–1541. - PubMed
-
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
