ATG16L1 autophagy pathway regulates BAX protein levels and programmed cell death
- PMID: 32848017
- PMCID: PMC7606669
- DOI: 10.1074/jbc.RA120.013999
ATG16L1 autophagy pathway regulates BAX protein levels and programmed cell death
Abstract
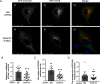
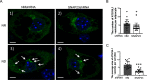
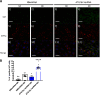
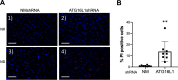
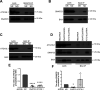
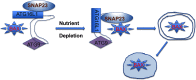
Previously we reported that adipocyte SNAP23 (synaptosome-associated protein of 23 kDa) deficiency blocks the activation of macroautophagy, leading to an increased abundance of BAX, a pro-death Bcl-2 family member, and activation and adipocyte cell death both in vitro and in vivo Here, we found that knockdown of SNAP23 inhibited the association of the autophagosome regulators ATG16L1 and ATG9 compartments by nutrient depletion and reduced the formation of ATG16L1 membrane puncta. ATG16L1 knockdown inhibited autophagy flux and increased BAX protein levels by suppressing BAX degradation. The elevation in BAX protein had no effect on BAX activation or cell death in the nutrient-replete state. However, following nutrient depletion, BAX was activated with a concomitant induction of cell death. Co-immunoprecipitation analyses demonstrated that SNAP23 and ATG16L1 proteins form a stable complex independent of nutrient condition, whereas in the nutrient-depleted state, BAX binds to SNAP23 to form a ternary BAX-SNAP23-ATG16L1 protein complex. Taken together, these data support a model in which SNAP23 plays a crucial function as a scaffold for ATG16L1 necessary for the suppression of BAX activation and induction of the intrinsic cell death program.
Keywords: ATG16L1; ATG9; BAX; SNAP23; SNARE proteins; adipocyte; apoptosis; autophagy; cell death; programmed.
© 2020 Chen et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
SNAP23 regulates BAX-dependent adipocyte programmed cell death independently of canonical macroautophagy.J Clin Invest. 2018 Aug 31;128(9):3941-3956. doi: 10.1172/JCI99217. Epub 2018 Aug 13. J Clin Invest. 2018. PMID: 30102258 Free PMC article.
-
Pancreas-specific SNAP23 depletion prevents pancreatitis by attenuating pathological basolateral exocytosis and formation of trypsin-activating autolysosomes.Autophagy. 2021 Oct;17(10):3068-3081. doi: 10.1080/15548627.2020.1852725. Epub 2020 Dec 7. Autophagy. 2021. PMID: 33213278 Free PMC article.
-
SNAP23 Regulates Endothelial Exocytosis of von Willebrand Factor.PLoS One. 2015 Aug 12;10(8):e0118737. doi: 10.1371/journal.pone.0118737. eCollection 2015. PLoS One. 2015. PMID: 26266817 Free PMC article.
-
HS1BP3 inhibits autophagy by regulation of PLD1.Autophagy. 2017 May 4;13(5):985-986. doi: 10.1080/15548627.2017.1291483. Epub 2017 Feb 25. Autophagy. 2017. PMID: 28318354 Free PMC article. Review.
-
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
Cited by
-
Autophagy Activation by Hypoxia Regulates Angiogenesis and Apoptosis in Oxidized Low-Density Lipoprotein-Induced Preeclampsia.Front Mol Biosci. 2021 Sep 9;8:709751. doi: 10.3389/fmolb.2021.709751. eCollection 2021. Front Mol Biosci. 2021. PMID: 34568425 Free PMC article.
-
Entamoeba histolytica exploits the autophagy pathway in macrophages to trigger inflammation in disease pathogenesis.Mucosal Immunol. 2021 Sep;14(5):1038-1054. doi: 10.1038/s41385-021-00408-4. Epub 2021 May 7. Mucosal Immunol. 2021. PMID: 33963264
-
Quercetin induces autophagy-associated death in HL-60 cells through CaMKKβ/AMPK/mTOR signal pathway.Acta Biochim Biophys Sin (Shanghai). 2022 Sep 25;54(9):1244-1256. doi: 10.3724/abbs.2022117. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 36148953 Free PMC article.
-
The Intestinal Microbiota May Be a Potential Theranostic Tool for Personalized Medicine.J Pers Med. 2022 Mar 24;12(4):523. doi: 10.3390/jpm12040523. J Pers Med. 2022. PMID: 35455639 Free PMC article. Review.
-
TFEB Biology and Agonists at a Glance.Cells. 2021 Feb 5;10(2):333. doi: 10.3390/cells10020333. Cells. 2021. PMID: 33562649 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

