Pentaminomycins C-E: Cyclic Pentapeptides as Autophagy Inducers from a Mealworm Beetle Gut Bacterium
- PMID: 32927831
- PMCID: PMC7565604
- DOI: 10.3390/microorganisms8091390
Pentaminomycins C-E: Cyclic Pentapeptides as Autophagy Inducers from a Mealworm Beetle Gut Bacterium
Abstract
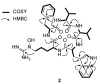
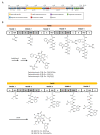
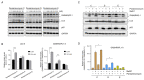
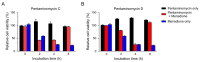
Pentaminomycins C-E (1-3) were isolated from the culture of the Streptomyces sp. GG23 strain from the guts of the mealworm beetle, Tenebrio molitor. The structures of the pentaminomycins were determined to be cyclic pentapeptides containing a modified amino acid, N5-hydroxyarginine, based on 1D and 2D NMR and mass spectroscopic analyses. The absolute configurations of the amino acid residues were assigned using Marfey's method and bioinformatics analysis of their nonribosomal peptide biosynthetic gene cluster (BGC). Detailed analysis of the BGC enabled us to propose that the structural variations in 1-3 originate from the low specificity of the adenylation domain in the nonribosomal peptide synthetase (NRPS) module 1, and indicate that macrocyclization can be catalyzed noncanonically by penicillin binding protein (PBP)-type TE. Furthermore, pentaminomycins C and D (1 and 2) showed significant autophagy-inducing activities and were cytoprotective against oxidative stress in vitro.
Keywords: OSMAC; autophagy inducer; biosynthetic pathway; cyclic peptides; gut bacteria; insect; mealworm.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Pentaminomycins F and G, Nonribosomal Peptides Containing 2-Pyridylalanine.J Nat Prod. 2021 Apr 23;84(4):1127-1134. doi: 10.1021/acs.jnatprod.0c01199. Epub 2021 Apr 1. J Nat Prod. 2021. PMID: 33793225
-
Pentaminomycins A and B, Hydroxyarginine-Containing Cyclic Pentapeptides from Streptomyces sp. RK88-1441.J Nat Prod. 2018 Apr 27;81(4):806-810. doi: 10.1021/acs.jnatprod.7b00882. Epub 2018 Mar 7. J Nat Prod. 2018. PMID: 29513529
-
One Pathway, Two Cyclic Non-Ribosomal Pentapeptides: Heterologous Expression of BE-18257 Antibiotics and Pentaminomycins from Streptomyces cacaoi CA-170360.Microorganisms. 2021 Jan 8;9(1):135. doi: 10.3390/microorganisms9010135. Microorganisms. 2021. PMID: 33430167 Free PMC article.
-
Nyuzenamide C, an Antiangiogenic Epoxy Cinnamic Acid-Containing Bicyclic Peptide from a Riverine Streptomyces sp.J Nat Prod. 2022 Apr 22;85(4):804-814. doi: 10.1021/acs.jnatprod.1c00837. Epub 2022 Mar 16. J Nat Prod. 2022. PMID: 35294831 Review.
-
Structural basis of the nonribosomal codes for nonproteinogenic amino acid selective adenylation enzymes in the biosynthesis of natural products.J Ind Microbiol Biotechnol. 2019 Mar;46(3-4):515-536. doi: 10.1007/s10295-018-2084-7. Epub 2018 Oct 5. J Ind Microbiol Biotechnol. 2019. PMID: 30291534 Review.
Cited by
-
Actinomycetes Associated with Arthropods as a Source of New Bioactive Compounds.Curr Issues Mol Biol. 2024 Apr 24;46(5):3822-3838. doi: 10.3390/cimb46050238. Curr Issues Mol Biol. 2024. PMID: 38785506 Free PMC article. Review.
-
Discovery and structure elucidation of glycosyl and 5-hydroxy migrastatins from dung beetle gut Kitasatospora sp.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuad046. doi: 10.1093/jimb/kuad046. J Ind Microbiol Biotechnol. 2023. PMID: 38093455 Free PMC article.
-
Microbial Exudates as Biostimulants: Role in Plant Growth Promotion and Stress Mitigation.J Xenobiot. 2023 Oct 1;13(4):572-603. doi: 10.3390/jox13040037. J Xenobiot. 2023. PMID: 37873814 Free PMC article. Review.
-
Antibiotics from Insect-Associated Actinobacteria.Biology (Basel). 2022 Nov 18;11(11):1676. doi: 10.3390/biology11111676. Biology (Basel). 2022. PMID: 36421390 Free PMC article. Review.
-
Harnessing phosphonate antibiotics argolaphos biosynthesis enables a synthetic biology-based green synthesis of glyphosate.Nat Commun. 2022 Apr 1;13(1):1736. doi: 10.1038/s41467-022-29188-6. Nat Commun. 2022. PMID: 35365617 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

