Virulence phenotypes result from interactions between pathogen ploidy and genetic background
- PMID: 32953064
- PMCID: PMC7487253
- DOI: 10.1002/ece3.6619
Virulence phenotypes result from interactions between pathogen ploidy and genetic background
Abstract
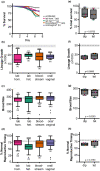
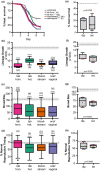
Studying fungal virulence is often challenging and frequently depends on many contexts, including host immune status and pathogen genetic background. However, the role of ploidy has often been overlooked when studying virulence in eukaryotic pathogens. Since fungal pathogens, including the human opportunistic pathogen Candida albicans, can display extensive ploidy variation, assessing how ploidy impacts virulence has important clinical relevance. As an opportunistic pathogen, C. albicans causes nonlethal, superficial infections in healthy individuals, but life-threatening bloodstream infections in individuals with compromised immune function. Here, we determined how both ploidy and genetic background of C. albicans impacts virulence phenotypes in healthy and immunocompromised nematode hosts by characterizing virulence phenotypes in four near-isogenic diploid and tetraploid pairs of strains, which included both laboratory and clinical genetic backgrounds. We found that C. albicans infections decreased host survival and negatively impacted host reproduction, and we leveraged these two measures to survey both lethal and nonlethal virulence phenotypes across the multiple C. albicans strains. In this study, we found that regardless of pathogen ploidy or genetic background, immunocompromised hosts were susceptible to fungal infection compared to healthy hosts. Furthermore, for each host context, we found a significant interaction between C. albicans genetic background and ploidy on virulence phenotypes, but no global differences between diploid and tetraploid pathogens were observed.
Keywords: fungi; pathogen; ploidy; virulence.
© 2020 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The author(s) declare no competing interests.
Figures



Similar articles
-
Increased Virulence and Large-Scale Reduction in Genome Size of Tetraploid Candida albicans Evolved in Nematode Hosts.Front Fungal Biol. 2022 Jun 27;3:903135. doi: 10.3389/ffunb.2022.903135. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746173 Free PMC article.
-
A Novel Virulence Phenotype Rapidly Assesses Candida Fungal Pathogenesis in Healthy and Immunocompromised Caenorhabditis elegans Hosts.mSphere. 2019 Apr 10;4(2):e00697-18. doi: 10.1128/mSphere.00697-18. mSphere. 2019. PMID: 30971447 Free PMC article.
-
Host-Induced Genome Instability Rapidly Generates Phenotypic Variation across Candida albicans Strains and Ploidy States.mSphere. 2020 Jun 3;5(3):e00433-20. doi: 10.1128/mSphere.00433-20. mSphere. 2020. PMID: 32493724 Free PMC article.
-
Genome plasticity in Candida albicans: A cutting-edge strategy for evolution, adaptation, and survival.Infect Genet Evol. 2022 Apr;99:105256. doi: 10.1016/j.meegid.2022.105256. Epub 2022 Feb 26. Infect Genet Evol. 2022. PMID: 35231665 Review.
-
Dissecting Candida albicans Infection from the Perspective of C. albicans Virulence and Omics Approaches on Host-Pathogen Interaction: A Review.Int J Mol Sci. 2016 Oct 18;17(10):1643. doi: 10.3390/ijms17101643. Int J Mol Sci. 2016. PMID: 27763544 Free PMC article. Review.
Cited by
-
Host Defense Mechanisms Induce Genome Instability Leading to Rapid Evolution in an Opportunistic Fungal Pathogen.Infect Immun. 2022 Feb 17;90(2):e0032821. doi: 10.1128/IAI.00328-21. Epub 2021 Dec 13. Infect Immun. 2022. PMID: 34898249 Free PMC article.
-
Increased Virulence and Large-Scale Reduction in Genome Size of Tetraploid Candida albicans Evolved in Nematode Hosts.Front Fungal Biol. 2022 Jun 27;3:903135. doi: 10.3389/ffunb.2022.903135. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746173 Free PMC article.
References
-
- Abbey, D. A. , Funt, J. , Lurie‐Weinberger, M. N. , Thompson, D. A. , Regev, A. , Myers, C. L. , & Berman, J. (2014). YMAP: A pipeline for visualization of copy number variation and loss of heterozygosity in eukaryotic pathogens. Genome Medicine, 6, 100 10.1186/PREACCEPT-1207699561372700 - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Research Materials

