Addressing the Possibility of a Histone-Like Code in Bacteria
- PMID: 32962352
- PMCID: PMC8177556
- DOI: 10.1021/acs.jproteome.0c00442
Addressing the Possibility of a Histone-Like Code in Bacteria
Abstract
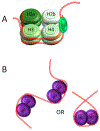
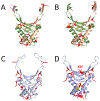
Acetylation was initially discovered as a post-translational modification (PTM) on the unstructured, highly basic N-terminal tails of eukaryotic histones in the 1960s. Histone acetylation constitutes part of the "histone code", which regulates chromosome compaction and various DNA processes such as gene expression, recombination, and DNA replication. In bacteria, nucleoid-associated proteins (NAPs) are responsible these functions in that they organize and compact the chromosome and regulate some DNA processes. The highly conserved DNABII family of proteins are considered functional homologues of eukaryotic histones despite having no sequence or structural conservation. Within the past decade, a growing interest in Nε-lysine acetylation led to the discovery that hundreds of bacterial proteins are acetylated with diverse cellular functions, in direct contrast to the original thought that this was a rare phenomenon. Similarly, other previously undiscovered bacterial PTMs, like serine, threonine, and tyrosine phosphorylation, have also been characterized. In this review, the various PTMs that were discovered among DNABII family proteins, specifically histone-like protein (HU) orthologues, from large-scale proteomic studies are discussed. The functional significance of these modifications and the enzymes involved are also addressed. The discovery of novel PTMs on these proteins begs this question: is there a histone-like code in bacteria?
Keywords: HBsu; HU; HupA; HupB; acetylation; acetyltransferase; deacetylase; phosphorylation; post-translational modification; succinylation.
Conflict of interest statement
The author declares no competing financial interest.
Figures
Similar articles
-
Nε-lysine acetylation of the histone-like protein HBsu influences antibiotic survival and persistence in Bacillus subtilis.Front Microbiol. 2024 May 21;15:1356733. doi: 10.3389/fmicb.2024.1356733. eCollection 2024. Front Microbiol. 2024. PMID: 38835483 Free PMC article.
-
YfmK is an Nε-lysine acetyltransferase that directly acetylates the histone-like protein HBsu in Bacillus subtilis.Proc Natl Acad Sci U S A. 2019 Feb 26;116(9):3752-3757. doi: 10.1073/pnas.1815511116. Epub 2019 Feb 11. Proc Natl Acad Sci U S A. 2019. PMID: 30808761 Free PMC article.
-
Integrative Chemical Biology Approaches to Deciphering the Histone Code: A Problem-Driven Journey.Acc Chem Res. 2021 Oct 5;54(19):3734-3747. doi: 10.1021/acs.accounts.1c00463. Epub 2021 Sep 23. Acc Chem Res. 2021. PMID: 34553920 Review.
-
Rv0802c is an acyltransferase that succinylates and acetylates Mycobacterium tuberculosis nucleoid-associated protein HU.Microbiology (Reading). 2021 Jul;167(7). doi: 10.1099/mic.0.001058. Microbiology (Reading). 2021. PMID: 34224344
-
Combinations of histone post-translational modifications.Biochem J. 2021 Feb 12;478(3):511-532. doi: 10.1042/BCJ20200170. Biochem J. 2021. PMID: 33567070 Review.
Cited by
-
Nε-Lysine Acetylation of the Histone-Like Protein HBsu Regulates the Process of Sporulation and Affects the Resistance Properties of Bacillus subtilis Spores.Front Microbiol. 2022 Jan 17;12:782815. doi: 10.3389/fmicb.2021.782815. eCollection 2021. Front Microbiol. 2022. PMID: 35111139 Free PMC article.
-
Nucleoid-associated proteins shape chromatin structure and transcriptional regulation across the bacterial kingdom.Transcription. 2021 Aug;12(4):182-218. doi: 10.1080/21541264.2021.1973865. Epub 2021 Sep 9. Transcription. 2021. PMID: 34499567 Free PMC article. Review.
-
Bacterial methyltransferases: from _targeting bacterial genomes to host epigenetics.Microlife. 2022 Aug 10;3:uqac014. doi: 10.1093/femsml/uqac014. eCollection 2022. Microlife. 2022. PMID: 37223361 Free PMC article. Review.
-
Nε-lysine acetylation of the histone-like protein HBsu influences antibiotic survival and persistence in Bacillus subtilis.Front Microbiol. 2024 May 21;15:1356733. doi: 10.3389/fmicb.2024.1356733. eCollection 2024. Front Microbiol. 2024. PMID: 38835483 Free PMC article.
-
Epigenetic-Mediated Antimicrobial Resistance: Host versus Pathogen Epigenetic Alterations.Antibiotics (Basel). 2022 Jun 16;11(6):809. doi: 10.3390/antibiotics11060809. Antibiotics (Basel). 2022. PMID: 35740215 Free PMC article. Review.
References
-
- Kornberg RD; Lorch Y Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999, 98 (3), 285–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

