COVID-19 therapy: What weapons do we bring into battle?
- PMID: 32992245
- PMCID: PMC7481143
- DOI: 10.1016/j.bmc.2020.115757
COVID-19 therapy: What weapons do we bring into battle?
Abstract
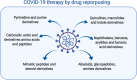
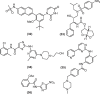
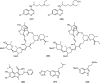
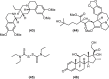
Urgent treatments, in any modality, to fight SARS-CoV-2 infections are desired by society in general, by health professionals, by Estate-leaders and, mainly, by the scientific community, because one thing is certain amidst the numerous uncertainties regarding COVID-19: knowledge is the means to discover or to produce an effective treatment against this global disease. Scientists from several areas in the world are still committed to this mission, as shown by the accelerated scientific production in the first half of 2020 with over 25,000 published articles related to the new coronavirus. Three great lines of publications related to COVID-19 were identified for building this article: The first refers to knowledge production concerning the virus and pathophysiology of COVID-19; the second regards efforts to produce vaccines against SARS-CoV-2 at a speed without precedent in the history of science; the third comprehends the attempts to find a marketed drug that can be used to treat COVID-19 by drug repurposing. In this review, the drugs that have been repurposed so far are grouped according to their chemical class. Their structures will be presented to provide better understanding of their structural similarities and possible correlations with mechanisms of actions. This can help identifying anti-SARS-CoV-2 promising therapeutic agents.
Keywords: Antiviral; COVID-19; Coronavirus; Repurposed drugs; SARS-CoV-2.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
COVID-19: Underpinning Research for Detection, Therapeutics, and Vaccines Development.Pharm Nanotechnol. 2020;8(4):323-353. doi: 10.2174/2211738508999200817163335. Pharm Nanotechnol. 2020. PMID: 32811406 Review.
-
A global picture: therapeutic perspectives for COVID-19.Immunotherapy. 2022 Apr;14(5):351-371. doi: 10.2217/imt-2021-0168. Epub 2022 Feb 21. Immunotherapy. 2022. PMID: 35187954 Free PMC article. Review.
-
COVID-19 Drug Development.J Microbiol Biotechnol. 2022 Jan 28;32(1):1-5. doi: 10.4014/jmb.2110.10029. J Microbiol Biotechnol. 2022. PMID: 34866128 Free PMC article. Review.
-
Potential Repurposed Therapeutics and New Vaccines against COVID-19 and Their Clinical Status.SLAS Discov. 2020 Dec;25(10):1097-1107. doi: 10.1177/2472555220945281. Epub 2020 Jul 21. SLAS Discov. 2020. PMID: 32692266 Free PMC article.
-
A Multidisciplinary Approach to Coronavirus Disease (COVID-19).Molecules. 2021 Jun 9;26(12):3526. doi: 10.3390/molecules26123526. Molecules. 2021. PMID: 34207756 Free PMC article. Review.
Cited by
-
Favipiravir and Its Structural Analogs: Antiviral Activity and Synthesis Methods.Acta Naturae. 2022 Apr-Jun;14(2):16-38. doi: 10.32607/actanaturae.11652. Acta Naturae. 2022. PMID: 35923566 Free PMC article.
-
Study to compare the effect of casirivimab and imdevimab, remdesivir, and favipiravir on progression and multi-organ function of hospitalized COVID-19 patients.Open Med (Wars). 2023 Aug 9;18(1):20230768. doi: 10.1515/med-2023-0768. eCollection 2023. Open Med (Wars). 2023. Retraction in: Open Med (Wars). 2023 Nov 10;18(1):20230853. doi: 10.1515/med-2023-0853 PMID: 37588660 Free PMC article. Retracted.
-
Synthesis, Antibacterial and Anthelmintic Activity of Novel 3-(3-Pyridyl)-oxazolidinone-5-methyl Ester Derivatives.Molecules. 2022 Feb 7;27(3):1103. doi: 10.3390/molecules27031103. Molecules. 2022. PMID: 35164368 Free PMC article.
-
Derivatives of benzo-1,4-thiazine-3-carboxylic acid and the corresponding amino acid conjugates.Beilstein J Org Chem. 2022 Sep 9;18:1195-1202. doi: 10.3762/bjoc.18.124. eCollection 2022. Beilstein J Org Chem. 2022. PMID: 36128428 Free PMC article.
-
Potency, Safety, and Pharmacokinetic Profiles of Potential Inhibitors _targeting SARS-CoV-2 Main Protease.Front Pharmacol. 2021 Feb 1;11:630500. doi: 10.3389/fphar.2020.630500. eCollection 2020. Front Pharmacol. 2021. PMID: 33597888 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

