Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma
- PMID: 33016930
- PMCID: PMC7598076
- DOI: 10.1172/JCI134132
Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma
Abstract
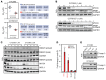
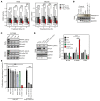
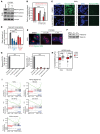
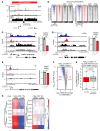
The undruggable nature of oncogenic Myc transcription factors poses a therapeutic challenge in neuroblastoma, a pediatric cancer in which MYCN amplification is strongly associated with unfavorable outcome. Here, we show that CYC065 (fadraciclib), a clinical inhibitor of CDK9 and CDK2, selectively _targeted MYCN-amplified neuroblastoma via multiple mechanisms. CDK9 - a component of the transcription elongation complex P-TEFb - bound to the MYCN-amplicon superenhancer, and its inhibition resulted in selective loss of nascent MYCN transcription. MYCN loss led to growth arrest, sensitizing cells for apoptosis following CDK2 inhibition. In MYCN-amplified neuroblastoma, MYCN invaded active enhancers, driving a transcriptionally encoded adrenergic gene expression program that was selectively reversed by CYC065. MYCN overexpression in mesenchymal neuroblastoma was sufficient to induce adrenergic identity and sensitize cells to CYC065. CYC065, used together with temozolomide, a reference therapy for relapsed neuroblastoma, caused long-term suppression of neuroblastoma growth in vivo, highlighting the clinical potential of CDK9/2 inhibition in the treatment of MYCN-amplified neuroblastoma.
Keywords: Cancer; Oncology; Transcription; Translation.
Conflict of interest statement
Figures

Similar articles
-
CDK4 inhibition restores G(1)-S arrest in MYCN-amplified neuroblastoma cells in the context of doxorubicin-induced DNA damage.Cell Cycle. 2013 Apr 1;12(7):1091-104. doi: 10.4161/cc.24091. Epub 2013 Mar 5. Cell Cycle. 2013. PMID: 23462184 Free PMC article.
-
Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells.Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12968-73. doi: 10.1073/pnas.0901418106. Epub 2009 Jun 12. Proc Natl Acad Sci U S A. 2009. PMID: 19525400 Free PMC article.
-
Combination therapy with the CDK7 inhibitor and the tyrosine kinase inhibitor exerts synergistic anticancer effects against MYCN-amplified neuroblastoma.Int J Cancer. 2020 Oct 1;147(7):1928-1938. doi: 10.1002/ijc.32936. Epub 2020 Mar 16. Int J Cancer. 2020. PMID: 32086952
-
Neuroblastoma-derived v-myc avian myelocytomatosis viral related oncogene or MYCN gene.J Clin Pathol. 2023 Aug;76(8):518-523. doi: 10.1136/jcp-2022-208476. Epub 2023 May 23. J Clin Pathol. 2023. PMID: 37221048 Review.
-
Didymin: an orally active citrus flavonoid for _targeting neuroblastoma.Onco_target. 2017 Apr 25;8(17):29428-29441. doi: 10.18632/onco_target.15204. Onco_target. 2017. PMID: 28187004 Free PMC article. Review.
Cited by
-
_targeting the Heterogeneous Genomic Landscape in Triple-Negative Breast Cancer through Inhibitors of the Transcriptional Machinery.Cancers (Basel). 2022 Sep 7;14(18):4353. doi: 10.3390/cancers14184353. Cancers (Basel). 2022. PMID: 36139513 Free PMC article. Review.
-
CDK9 inhibitors for the treatment of solid tumors.Biochem Pharmacol. 2024 Nov;229:116470. doi: 10.1016/j.bcp.2024.116470. Epub 2024 Aug 8. Biochem Pharmacol. 2024. PMID: 39127153 Review.
-
_targeting cyclin-dependent kinase 9 in cancer therapy.Acta Pharmacol Sin. 2022 Jul;43(7):1633-1645. doi: 10.1038/s41401-021-00796-0. Epub 2021 Nov 22. Acta Pharmacol Sin. 2022. PMID: 34811514 Free PMC article. Review.
-
Transcriptional CDK inhibitors, CYC065 and THZ1 promote Bim-dependent apoptosis in primary and recurrent GBM through cell cycle arrest and Mcl-1 downregulation.Cell Death Dis. 2021 Aug 3;12(8):763. doi: 10.1038/s41419-021-04050-7. Cell Death Dis. 2021. PMID: 34344865 Free PMC article.
-
_targeting CDK9 with selective inhibitors or degraders in tumor therapy: an overview of recent developments.Cancer Biol Ther. 2023 Dec 31;24(1):2219470. doi: 10.1080/15384047.2023.2219470. Cancer Biol Ther. 2023. PMID: 37272701 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- 22897/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_18051/MRC_/Medical Research Council/United Kingdom
- 091763Z/10/Z/WT_/Wellcome Trust/United Kingdom
- C34648/A28278/CRUK_/Cancer Research UK/United Kingdom
- C1060/A10334/CRUK_/Cancer Research UK/United Kingdom
- C1090/A16464/CRUK_/Cancer Research UK/United Kingdom
- 27725/CRUK_/Cancer Research UK/United Kingdom
- C34648/A18339/CRUK_/Cancer Research UK/United Kingdom
- 16464/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_16047/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 14610/CRUK_/Cancer Research UK/United Kingdom
- R01 CA215452/CA/NCI NIH HHS/United States
- C309/A11566/CRUK_/Cancer Research UK/United Kingdom
- 23302/CRUK_/Cancer Research UK/United Kingdom
- 28278/CRUK_/Cancer Research UK/United Kingdom
- C24461/A23302/CRUK_/Cancer Research UK/United Kingdom
- DH_/Department of Health/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

