Virome of Bat Guano from Nine Northern California Roosts
- PMID: 33115864
- PMCID: PMC7925108
- DOI: 10.1128/JVI.01713-20
Virome of Bat Guano from Nine Northern California Roosts
Abstract
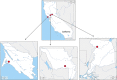
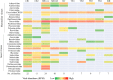
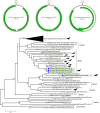
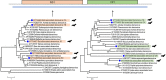
Bats are hosts to a large variety of viruses, including many capable of cross-species transmissions to other mammals, including humans. We characterized the virome in guano from five common bat species in 9 Northern California roosts and from a pool of 5 individual bats. Genomes belonging to 14 viral families known to infect mammals and 17 viral families infecting insects or of unknown tropism were detected. Nearly complete or complete genomes of a novel parvovirus, astrovirus, nodavirus, circular Rep-encoding single-stranded DNA (CRESS-DNA) viruses, and densoviruses, and more partial genomes of a novel alphacoronavirus and a bunyavirus were characterized. Lower numbers of reads with >90% amino acid identity to previously described calicivirus, circovirus, adenoviruses, hepatovirus, bocaparvoviruses, and polyomavirus in other bat species were also found, likely reflecting their wide distribution among different bats. Unexpectedly, a few sequence reads of canine parvovirus 2 and the recently described mouse kidney parvovirus were also detected and their presence confirmed by PCR; these possibly originated from guano contamination by carnivores and rodents. The majority of eukaryotic viral reads were highly divergent, indicating that numerous viruses still remain to be characterized, even from such a heavily investigated order as Chiroptera.IMPORTANCE Characterizing the bat virome is important for understanding viral diversity and detecting viral spillover between animal species. Using an unbiased metagenomics method, we characterize the virome in guano collected from multiple roosts of common Northern California bat species. We describe several novel viral genomes and report the detection of viruses with close relatives reported in other bat species, likely reflecting cross-species transmissions. Viral sequences from well-known carnivore and rodent parvoviruses were also detected, whose presence are likely the result of contamination from defecation and urination atop guano and which reflect the close interaction of these mammals in the wild.
Keywords: bat virome; emerging viruses; metagenomics.
Copyright © 2021 American Society for Microbiology.
Figures




Similar articles
-
Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses.J Virol. 2010 Jul;84(14):6955-65. doi: 10.1128/JVI.00501-10. Epub 2010 May 12. J Virol. 2010. PMID: 20463061 Free PMC article.
-
The Myotis chiloensis Guano Virome: Viral Nucleic Acid Enrichments for High-Resolution Virome Elucidation and Full Alphacoronavirus Genome Assembly.Viruses. 2022 Jan 20;14(2):202. doi: 10.3390/v14020202. Viruses. 2022. PMID: 35215796 Free PMC article.
-
Viral Metagenomic Profiling of Croatian Bat Population Reveals Sample and Habitat Dependent Diversity.Viruses. 2020 Aug 14;12(8):891. doi: 10.3390/v12080891. Viruses. 2020. PMID: 32824037 Free PMC article.
-
The Virome of Bats Inhabiting Brazilian Biomes: Knowledge Gaps and Biases towards Zoonotic Viruses.Microbiol Spectr. 2023 Feb 14;11(1):e0407722. doi: 10.1128/spectrum.04077-22. Epub 2023 Jan 10. Microbiol Spectr. 2023. PMID: 36625641 Free PMC article. Review.
-
Bat virome research: the past, the present and the future.Curr Opin Virol. 2021 Aug;49:68-80. doi: 10.1016/j.coviro.2021.04.013. Epub 2021 May 27. Curr Opin Virol. 2021. PMID: 34052731 Review.
Cited by
-
Characterization of Pipistrellus pygmaeus Bat Virome from Sweden.Viruses. 2022 Jul 28;14(8):1654. doi: 10.3390/v14081654. Viruses. 2022. PMID: 36016275 Free PMC article.
-
Novel Virus Identification through Metagenomics: A Systematic Review.Life (Basel). 2022 Dec 7;12(12):2048. doi: 10.3390/life12122048. Life (Basel). 2022. PMID: 36556413 Free PMC article. Review.
-
Plasma Virome Reveals Blooms and Transmission of Anellovirus in Intravenous Drug Users with HIV-1, HCV, and/or HBV Infections.Microbiol Spectr. 2022 Aug 31;10(4):e0144722. doi: 10.1128/spectrum.01447-22. Epub 2022 Jun 27. Microbiol Spectr. 2022. PMID: 35758682 Free PMC article.
-
Myotis fimbriatus Virome, a Window to Virus Diversity and Evolution in the Genus Myotis.Viruses. 2022 Aug 27;14(9):1899. doi: 10.3390/v14091899. Viruses. 2022. PMID: 36146706 Free PMC article.
-
Phylogenetic Diversity of Animal Oral and Gastrointestinal Viromes Useful in Surveillance of Zoonoses.Microorganisms. 2022 Sep 10;10(9):1815. doi: 10.3390/microorganisms10091815. Microorganisms. 2022. PMID: 36144417 Free PMC article. Review.
References
-
- Fenton M, Simmons N. 2015. Bats: a world of science and mystery, 1st ed. University of Chicago Press, Chicago, IL.
-
- Bolatti EM, Zorec TM, Montani ME, Hošnjak L, Chouhy D, Viarengo G, Casal PE, Barquez RM, Poljak M, Giri AA. 2020. A preliminary study of the virome of the South American free-tailed bats (Tadarida brasiliensis) and identification of two novel mammalian viruses. Viruses 12:422. doi:10.3390/v12040422. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

