Focus on DNA Glycosylases-A Set of Tightly Regulated Enzymes with a High Potential as Anticancer Drug _targets
- PMID: 33287345
- PMCID: PMC7730500
- DOI: 10.3390/ijms21239226
Focus on DNA Glycosylases-A Set of Tightly Regulated Enzymes with a High Potential as Anticancer Drug _targets
Abstract
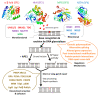
Cancer is the second leading cause of death with tens of millions of people diagnosed with cancer every year around the world. Most radio- and chemotherapies aim to eliminate cancer cells, notably by causing severe damage to the DNA. However, efficient repair of such damage represents a common mechanism of resistance to initially effective cytotoxic agents. Thus, development of new generation anticancer drugs that _target DNA repair pathways, and more particularly the base excision repair (BER) pathway that is responsible for removal of damaged bases, is of growing interest. The BER pathway is initiated by a set of enzymes known as DNA glycosylases. Unlike several downstream BER enzymes, DNA glycosylases have so far received little attention and the development of specific inhibitors of these enzymes has been lagging. Yet, dysregulation of DNA glycosylases is also known to play a central role in numerous cancers and at different stages of the disease, and thus inhibiting DNA glycosylases is now considered a valid strategy to eliminate cancer cells. This review provides a detailed overview of the activities of DNA glycosylases in normal and cancer cells, their modes of regulation, and their potential as anticancer drug _targets.
Keywords: DNA glycosylases; base excision repair; cancer; drug resistance; inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Human DNA glycosylases involved in the repair of oxidatively damaged DNA.Biol Pharm Bull. 2004 Apr;27(4):480-5. doi: 10.1248/bpb.27.480. Biol Pharm Bull. 2004. PMID: 15056851 Review.
-
Oxidative DNA damage repair in mammalian cells: a new perspective.DNA Repair (Amst). 2007 Apr 1;6(4):470-80. doi: 10.1016/j.dnarep.2006.10.011. Epub 2006 Nov 20. DNA Repair (Amst). 2007. PMID: 17116430 Free PMC article. Review.
-
Oxidative Damage in Sporadic Colorectal Cancer: Molecular Mapping of Base Excision Repair Glycosylases in Colorectal Cancer Patients.Int J Mol Sci. 2020 Apr 2;21(7):2473. doi: 10.3390/ijms21072473. Int J Mol Sci. 2020. PMID: 32252452 Free PMC article. Review.
-
Emerging Roles of DNA Glycosylases and the Base Excision Repair Pathway.Trends Biochem Sci. 2019 Sep;44(9):765-781. doi: 10.1016/j.tibs.2019.04.006. Epub 2019 May 9. Trends Biochem Sci. 2019. PMID: 31078398 Free PMC article. Review.
-
Small Molecule Inhibitors of 8-Oxoguanine DNA Glycosylase-1 (OGG1).ACS Chem Biol. 2015 Oct 16;10(10):2334-43. doi: 10.1021/acschembio.5b00452. Epub 2015 Aug 7. ACS Chem Biol. 2015. PMID: 26218629 Free PMC article.
Cited by
-
Detection of Uracil-Excising DNA Glycosylases in Cancer Cell Samples Using a Three-Dimensional DNAzyme Walker.ACS Meas Sci Au. 2024 May 8;4(4):459-466. doi: 10.1021/acsmeasuresciau.4c00011. eCollection 2024 Aug 21. ACS Meas Sci Au. 2024. PMID: 39184356 Free PMC article.
-
New Discoveries on Protein Recruitment and Regulation during the Early Stages of the DNA Damage Response Pathways.Int J Mol Sci. 2024 Jan 30;25(3):1676. doi: 10.3390/ijms25031676. Int J Mol Sci. 2024. PMID: 38338953 Free PMC article. Review.
-
TRIM26 Maintains Cell Survival in Response to Oxidative Stress through Regulating DNA Glycosylase Stability.Int J Mol Sci. 2022 Oct 1;23(19):11613. doi: 10.3390/ijms231911613. Int J Mol Sci. 2022. PMID: 36232914 Free PMC article.
-
Characterization of a Novel Thermostable DNA Lyase Used To Prepare DNA for Next-Generation Sequencing.Chem Res Toxicol. 2023 Feb 20;36(2):162-176. doi: 10.1021/acs.chemrestox.2c00172. Epub 2023 Jan 16. Chem Res Toxicol. 2023. PMID: 36647573 Free PMC article.
-
The H2TH-like motif of the Escherichia coli multifunctional protein KsgA is required for DNA binding involved in DNA repair and the suppression of mutation frequencies.Genes Environ. 2023 Apr 12;45(1):13. doi: 10.1186/s41021-023-00266-5. Genes Environ. 2023. PMID: 37041652 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

