Chromatin accessibility maps provide evidence of multilineage gene priming in hematopoietic stem cells
- PMID: 33407811
- PMCID: PMC7789351
- DOI: 10.1186/s13072-020-00377-1
Chromatin accessibility maps provide evidence of multilineage gene priming in hematopoietic stem cells
Abstract
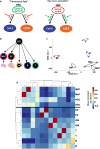
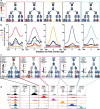
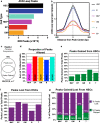
Hematopoietic stem cells (HSCs) have the capacity to differentiate into vastly different types of mature blood cells. The epigenetic mechanisms regulating the multilineage ability, or multipotency, of HSCs are not well understood. To test the hypothesis that cis-regulatory elements that control fate decisions for all lineages are primed in HSCs, we used ATAC-seq to compare chromatin accessibility of HSCs with five unipotent cell types. We observed the highest similarity in accessibility profiles between megakaryocyte progenitors and HSCs, whereas B cells had the greatest number of regions with de novo gain in accessibility during differentiation. Despite these differences, we identified cis-regulatory elements from all lineages that displayed epigenetic priming in HSCs. These findings provide new insights into the regulation of stem cell multipotency, as well as a resource to identify functional drivers of lineage fate.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Dynamics of Chromatin Accessibility During Hematopoietic Stem Cell Differentiation Into Progressively Lineage-Committed Progeny.Stem Cells. 2023 May 15;41(5):520-539. doi: 10.1093/stmcls/sxad022. Stem Cells. 2023. PMID: 36945732 Free PMC article.
-
Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points.Epigenetics Chromatin. 2018 May 28;11(1):22. doi: 10.1186/s13072-018-0195-z. Epigenetics Chromatin. 2018. PMID: 29807547 Free PMC article.
-
Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells.Nature. 2018 Feb 1;554(7690):106-111. doi: 10.1038/nature25455. Epub 2018 Jan 3. Nature. 2018. PMID: 29298288
-
[Hematopoietic Stem Cells Differentiate into the Megakaryocyte Lineage--Review].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020 Jun;28(3):1044-1048. doi: 10.19746/j.cnki.issn.1009-2137.2020.03.054. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2020. PMID: 32552979 Review. Chinese.
-
Altering chromatin methylation patterns and the transcriptional network involved in regulation of hematopoietic stem cell fate.J Cell Physiol. 2020 Oct;235(10):6404-6423. doi: 10.1002/jcp.29642. Epub 2020 Feb 13. J Cell Physiol. 2020. PMID: 32052445 Review.
Cited by
-
Single cell spatial biology over developmental time can decipher pediatric brain pathologies.Neurobiol Dis. 2024 Sep;199:106597. doi: 10.1016/j.nbd.2024.106597. Epub 2024 Jul 9. Neurobiol Dis. 2024. PMID: 38992777 Review.
-
Chromatin accessibility and cell cycle progression are controlled by the HDAC-associated Sin3B protein in murine hematopoietic stem cells.Epigenetics Chromatin. 2024 Jan 23;17(1):2. doi: 10.1186/s13072-024-00526-w. Epigenetics Chromatin. 2024. PMID: 38254205 Free PMC article.
-
Single-cell ATAC and RNA sequencing reveal pre-existing and persistent cells associated with prostate cancer relapse.Nat Commun. 2021 Sep 6;12(1):5307. doi: 10.1038/s41467-021-25624-1. Nat Commun. 2021. PMID: 34489465 Free PMC article.
-
EnhancerNet: a predictive model of cell identity dynamics through enhancer selection.Development. 2024 Oct 1;151(19):dev202997. doi: 10.1242/dev.202997. Epub 2024 Oct 9. Development. 2024. PMID: 39289870 Free PMC article.
-
A quantitative hematopoietic stem cell reconstitution protocol: Accounting for recipient variability, tissue distribution and cell half-lives.Stem Cell Res. 2020 Dec 29;50:102145. doi: 10.1016/j.scr.2020.102145. Online ahead of print. Stem Cell Res. 2020. PMID: 33486300 Free PMC article.
References
-
- Bender MA, Ragoczy T, Lee J, Byron R, Telling A, Dean A, Groudine M. The hypersensitive sites of the murine β-globin locus control region act independently to affect nuclear localization and transcriptional elongation. Blood. 2012;119:3820–3827. doi: 10.1182/blood-2011-09-380485. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

