SARS-CoV-2 evolution during treatment of chronic infection
- PMID: 33545711
- PMCID: PMC7610568
- DOI: 10.1038/s41586-021-03291-y
SARS-CoV-2 evolution during treatment of chronic infection
Erratum in
-
Author Correction: SARS-CoV-2 evolution during treatment of chronic infection.Nature. 2022 Aug;608(7922):E23. doi: 10.1038/s41586-022-05104-2. Nature. 2022. PMID: 35864233 Free PMC article. No abstract available.
Abstract
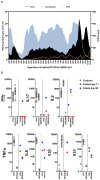
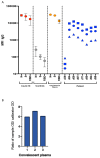
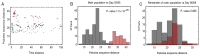
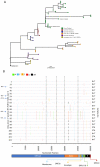
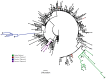
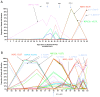
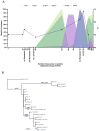
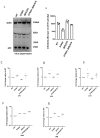
The spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is critical for virus infection through the engagement of the human ACE2 protein1 and is a major antibody _target. Here we show that chronic infection with SARS-CoV-2 leads to viral evolution and reduced sensitivity to neutralizing antibodies in an immunosuppressed individual treated with convalescent plasma, by generating whole-genome ultra-deep sequences for 23 time points that span 101 days and using in vitro techniques to characterize the mutations revealed by sequencing. There was little change in the overall structure of the viral population after two courses of remdesivir during the first 57 days. However, after convalescent plasma therapy, we observed large, dynamic shifts in the viral population, with the emergence of a dominant viral strain that contained a substitution (D796H) in the S2 subunit and a deletion (ΔH69/ΔV70) in the S1 N-terminal domain of the spike protein. As passively transferred serum antibodies diminished, viruses with the escape genotype were reduced in frequency, before returning during a final, unsuccessful course of convalescent plasma treatment. In vitro, the spike double mutant bearing both ΔH69/ΔV70 and D796H conferred modestly decreased sensitivity to convalescent plasma, while maintaining infectivity levels that were similar to the wild-type virus.The spike substitution mutant D796H appeared to be the main contributor to the decreased susceptibility to neutralizing antibodies, but this mutation resulted in an infectivity defect. The spike deletion mutant ΔH69/ΔV70 had a twofold higher level of infectivity than wild-type SARS-CoV-2, possibly compensating for the reduced infectivity of the D796H mutation. These data reveal strong selection on SARS-CoV-2 during convalescent plasma therapy, which is associated with the emergence of viral variants that show evidence of reduced susceptibility to neutralizing antibodies in immunosuppressed individuals.
Conflict of interest statement
Figures





Update of
-
Neutralising antibodies in Spike mediated SARS-CoV-2 adaptation.medRxiv [Preprint]. 2020 Dec 29:2020.12.05.20241927. doi: 10.1101/2020.12.05.20241927. medRxiv. 2020. Update in: Nature. 2021 Apr;592(7853):277-282. doi: 10.1038/s41586-021-03291-y PMID: 33398302 Free PMC article. Updated. Preprint.
Similar articles
-
Neutralising antibodies in Spike mediated SARS-CoV-2 adaptation.medRxiv [Preprint]. 2020 Dec 29:2020.12.05.20241927. doi: 10.1101/2020.12.05.20241927. medRxiv. 2020. Update in: Nature. 2021 Apr;592(7853):277-282. doi: 10.1038/s41586-021-03291-y PMID: 33398302 Free PMC article. Updated. Preprint.
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
Emergence of Multiple SARS-CoV-2 Antibody Escape Variants in an Immunocompromised Host Undergoing Convalescent Plasma Treatment.mSphere. 2021 Aug 25;6(4):e0048021. doi: 10.1128/mSphere.00480-21. Epub 2021 Aug 25. mSphere. 2021. PMID: 34431691 Free PMC article.
-
Neutralising antibody escape of SARS-CoV-2 spike protein: Risk assessment for antibody-based Covid-19 therapeutics and vaccines.Rev Med Virol. 2021 Nov;31(6):e2231. doi: 10.1002/rmv.2231. Epub 2021 Mar 16. Rev Med Virol. 2021. PMID: 33724631 Free PMC article. Review.
-
SARS-CoV-2 journey: from alpha variant to omicron and its sub-variants.Infection. 2024 Jun;52(3):767-786. doi: 10.1007/s15010-024-02223-y. Epub 2024 Mar 30. Infection. 2024. PMID: 38554253 Free PMC article. Review.
Cited by
-
Tackling COVID-19 with neutralizing monoclonal antibodies.Cell. 2021 Jun 10;184(12):3086-3108. doi: 10.1016/j.cell.2021.05.005. Epub 2021 May 26. Cell. 2021. PMID: 34087172 Free PMC article. Review.
-
SARS-CoV-2 Genetic Diversity and Lineage Dynamics in Egypt during the First 18 Months of the Pandemic.Viruses. 2022 Aug 25;14(9):1878. doi: 10.3390/v14091878. Viruses. 2022. PMID: 36146685 Free PMC article.
-
Rapid intra-host diversification and evolution of SARS-CoV-2 in advanced HIV infection.Nat Commun. 2024 Aug 22;15(1):7240. doi: 10.1038/s41467-024-51539-8. Nat Commun. 2024. PMID: 39174553 Free PMC article.
-
Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera.Cell. 2021 Apr 29;184(9):2348-2361.e6. doi: 10.1016/j.cell.2021.02.037. Epub 2021 Feb 23. Cell. 2021. PMID: 33730597 Free PMC article.
-
Evaluating the impact of COVID-19 on cancer declarations in Quebec, Canada.Cancer Med. 2023 Mar;12(5):6260-6269. doi: 10.1002/cam4.5389. Epub 2022 Nov 16. Cancer Med. 2023. PMID: 36385491 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- G0701652/MRC_/Medical Research Council/United Kingdom
- 108082/WT_/Wellcome Trust/United Kingdom
- 207498/WT_/Wellcome Trust/United Kingdom
- 206618/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MC_UP_1201/16/MRC_/Medical Research Council/United Kingdom
- MC_PC_19027/MRC_/Medical Research Council/United Kingdom
- 204911/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- R01 GM083127/GM/NIGMS NIH HHS/United States
- BB/P007562/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UU_00002/11/MRC_/Medical Research Council/United Kingdom
- MR/R008698/1/MRC_/Medical Research Council/United Kingdom
- RP-PG-0514-20018/DH_/Department of Health/United Kingdom
- MR/R015600/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

