Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera
- PMID: 33730597
- PMCID: PMC7901269
- DOI: 10.1016/j.cell.2021.02.037
Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera
Abstract
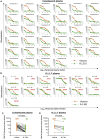
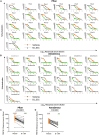
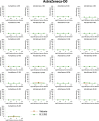
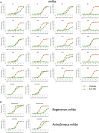
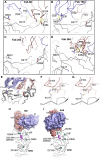
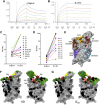
The race to produce vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) began when the first sequence was published, and this forms the basis for vaccines currently deployed globally. Independent lineages of SARS-CoV-2 have recently been reported: UK, B.1.1.7; South Africa, B.1.351; and Brazil, P.1. These variants have multiple changes in the immunodominant spike protein that facilitates viral cell entry via the angiotensin-converting enzyme-2 (ACE2) receptor. Mutations in the receptor recognition site on the spike are of great concern for their potential for immune escape. Here, we describe a structure-function analysis of B.1.351 using a large cohort of convalescent and vaccinee serum samples. The receptor-binding domain mutations provide tighter ACE2 binding and widespread escape from monoclonal antibody neutralization largely driven by E484K, although K417N and N501Y act together against some important antibody classes. In a number of cases, it would appear that convalescent and some vaccine serum offers limited protection against this variant.
Keywords: ACE2; B.1.351; SARS-CoV-2; South Africa; antibody; escape; neutralization; receptor-binding domain; vaccine; variant.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests G.R.S. sits on the GSK Vaccines Scientific Advisory Board. Oxford University holds intellectual property related to the Oxford-AstraZeneca vaccine. A.J.P. is Chair of UK Department Health and Social Care’s (DHSC) Joint Committee on Vaccination & Immunisation (JCVI), but does not participate in the JCVI COVID-19 committee, and is a member of the WHO’s SAGE. The views expressed in this article do not necessarily represent the views of DHSC, JCVI, or WHO. The University of Oxford has entered into a partnership with AstraZeneca on coronavirus vaccine development. The University of Oxford has protected intellectual property disclosed in this publication.
Figures


Similar articles
-
Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7.Nature. 2021 May;593(7857):130-135. doi: 10.1038/s41586-021-03398-2. Epub 2021 Mar 8. Nature. 2021. PMID: 33684923
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
Antibody evasion by the P.1 strain of SARS-CoV-2.Cell. 2021 May 27;184(11):2939-2954.e9. doi: 10.1016/j.cell.2021.03.055. Epub 2021 Mar 30. Cell. 2021. PMID: 33852911 Free PMC article.
-
Neutralising antibody escape of SARS-CoV-2 spike protein: Risk assessment for antibody-based Covid-19 therapeutics and vaccines.Rev Med Virol. 2021 Nov;31(6):e2231. doi: 10.1002/rmv.2231. Epub 2021 Mar 16. Rev Med Virol. 2021. PMID: 33724631 Free PMC article. Review.
-
Variants of SARS-CoV-2, their effects on infection, transmission and neutralization by vaccine-induced antibodies.Eur Rev Med Pharmacol Sci. 2021 Sep;25(18):5857-5864. doi: 10.26355/eurrev_202109_26805. Eur Rev Med Pharmacol Sci. 2021. PMID: 34604978 Review.
Cited by
-
Statistical Challenges in Tracking the Evolution of SARS-CoV-2.Stat Sci. 2022 May;37(2):162-182. doi: 10.1214/22-sts853. Epub 2022 May 16. Stat Sci. 2022. PMID: 36034090 Free PMC article.
-
mRNA vaccination compared to infection elicits an IgG-predominant response with greater SARS-CoV-2 specificity and similar decrease in variant spike recognition.medRxiv [Preprint]. 2021 Apr 7:2021.04.05.21254952. doi: 10.1101/2021.04.05.21254952. medRxiv. 2021. PMID: 33851181 Free PMC article. Preprint.
-
Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination.Sci Immunol. 2021 Apr 15;6(58):eabi6950. doi: 10.1126/sciimmunol.abi6950. Sci Immunol. 2021. PMID: 33858945 Free PMC article.
-
Surveillance for SARS-CoV-2 variants of concern in the Australian context.Med J Aust. 2021 Jun;214(11):500-502.e1. doi: 10.5694/mja2.51105. Epub 2021 May 31. Med J Aust. 2021. PMID: 34060079 Free PMC article. No abstract available.
-
SARS-CoV-2 mRNA vaccination elicits broad and potent Fc effector functions to VOCs in vulnerable populations.medRxiv [Preprint]. 2022 Sep 18:2022.09.15.22280000. doi: 10.1101/2022.09.15.22280000. medRxiv. 2022. Update in: Nat Commun. 2023 Aug 24;14(1):5171. doi: 10.1038/s41467-023-40960-0 PMID: 36172122 Free PMC article. Updated. Preprint.
References
-
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

