Dynamic Responses of Barley Root Succinyl-Proteome to Short-Term Phosphate Starvation and Recovery
- PMID: 33868348
- PMCID: PMC8045032
- DOI: 10.3389/fpls.2021.649147
Dynamic Responses of Barley Root Succinyl-Proteome to Short-Term Phosphate Starvation and Recovery
Abstract
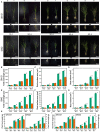
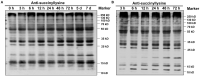
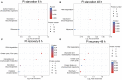
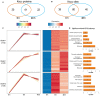
Barley (Hordeum vulgare L.)-a major cereal crop-has low Pi demand, which is a distinct advantage for studying the tolerance mechanisms of phosphorus deficiency. We surveyed dynamic protein succinylation events in barley roots in response to and recovery from Pi starvation by firstly evaluating the impact of Pi starvation in a Pi-tolerant (GN121) and Pi-sensitive (GN42) barley genotype exposed to long-term low Pi (40 d) followed by a high-Pi recovery for 10 d. An integrated proteomics approach involving label-free, immune-affinity enrichment, and high-resolution LC-MS/MS spectrometric analysis was then used to quantify succinylome and proteome in GN121 roots under short-term Pi starvation (6, 48 h) and Pi recovery (6, 48 h). We identified 2,840 succinylation sites (Ksuc) across 884 proteins; of which, 11 representative Ksuc motifs had the preferred amino acid residue (lysine). Furthermore, there were 81 differentially abundant succinylated proteins (DFASPs) from 119 succinylated sites, 83 DFASPs from 110 succinylated sites, 93 DFASPs from 139 succinylated sites, and 91 DFASPs from 123 succinylated sites during Pi starvation for 6 and 48 h and during Pi recovery for 6 and 48 h, respectively. Pi starvation enriched ribosome pathways, glycolysis, and RNA degradation. Pi recovery enriched the TCA cycle, glycolysis, and oxidative phosphorylation. Importantly, many of the DFASPs identified during Pi starvation were significantly overexpressed during Pi recovery. These results suggest that barley roots can regulate specific Ksuc site changes in response to Pi stress as well as specific metabolic processes. Resolving the metabolic pathways of succinylated protein regulation characteristics will improve phosphate acquisition and utilization efficiency in crops.
Keywords: Hordeum vulgare L.; Pi stress; germplasm; metabolism; succinylated protein.
Copyright © 2021 Wang, Ma, Li, Ren, Yao, Li, Meng, Ma, Si, Yang, Shang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Protein post-translational modification by lysine succinylation: Biochemistry, biological implications, and therapeutic opportunities.Genes Dis. 2022 Apr 7;10(4):1242-1262. doi: 10.1016/j.gendis.2022.03.009. eCollection 2023 Jul. Genes Dis. 2022. PMID: 37397549 Free PMC article. Review.
-
Global Profiling of Phosphorylation Reveals the Barley Roots Response to Phosphorus Starvation and Resupply.Front Plant Sci. 2021 Jul 14;12:676432. doi: 10.3389/fpls.2021.676432. eCollection 2021. Front Plant Sci. 2021. PMID: 34335649 Free PMC article.
-
Global proteome analyses of phosphorylation and succinylation of barley root proteins in response to phosphate starvation and recovery.Front Plant Sci. 2022 Aug 18;13:917652. doi: 10.3389/fpls.2022.917652. eCollection 2022. Front Plant Sci. 2022. PMID: 36061799 Free PMC article.
-
Molecular Mechanisms of Acclimatization to Phosphorus Starvation and Recovery Underlying Full-Length Transcriptome Profiling in Barley (Hordeum vulgare L.).Front Plant Sci. 2018 Apr 18;9:500. doi: 10.3389/fpls.2018.00500. eCollection 2018. Front Plant Sci. 2018. PMID: 29720989 Free PMC article.
-
Succinyl-proteome profiling of Dendrobium officinale, an important traditional Chinese orchid herb, revealed involvement of succinylation in the glycolysis pathway.BMC Genomics. 2017 Aug 10;18(1):598. doi: 10.1186/s12864-017-3978-x. BMC Genomics. 2017. PMID: 28797234 Free PMC article.
Cited by
-
Plant Proteoforms Under Environmental Stress: Functional Proteins Arising From a Single Gene.Front Plant Sci. 2021 Dec 14;12:793113. doi: 10.3389/fpls.2021.793113. eCollection 2021. Front Plant Sci. 2021. PMID: 34970290 Free PMC article. Review.
-
Exogenous Melatonin Enhances the Low Phosphorus Tolerance of Barley Roots of Different Genotypes.Cells. 2023 May 16;12(10):1397. doi: 10.3390/cells12101397. Cells. 2023. PMID: 37408231 Free PMC article.
-
Protein post-translational modification by lysine succinylation: Biochemistry, biological implications, and therapeutic opportunities.Genes Dis. 2022 Apr 7;10(4):1242-1262. doi: 10.1016/j.gendis.2022.03.009. eCollection 2023 Jul. Genes Dis. 2022. PMID: 37397549 Free PMC article. Review.
-
Global Profiling of Phosphorylation Reveals the Barley Roots Response to Phosphorus Starvation and Resupply.Front Plant Sci. 2021 Jul 14;12:676432. doi: 10.3389/fpls.2021.676432. eCollection 2021. Front Plant Sci. 2021. PMID: 34335649 Free PMC article.
-
Proteomic Analysis Dissects Molecular Mechanisms Underlying Plant Responses to Phosphorus Deficiency.Cells. 2022 Feb 14;11(4):651. doi: 10.3390/cells11040651. Cells. 2022. PMID: 35203302 Free PMC article. Review.
References
-
- Cordell D., Drangert J., White S. (2009). The story of phosphorus: global food security and food for thought. Glob. Environ. Change 19, 292–305. 10.1016/j.gloenvcha.2008.10.009 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

