Differential Transcriptomes and Methylomes of Trophoblast Stem Cells From Naturally-Fertilized and Somatic Cell Nuclear-Transferred Embryos
- PMID: 33869230
- PMCID: PMC8047118
- DOI: 10.3389/fcell.2021.664178
Differential Transcriptomes and Methylomes of Trophoblast Stem Cells From Naturally-Fertilized and Somatic Cell Nuclear-Transferred Embryos
Abstract
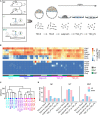
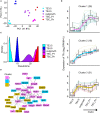
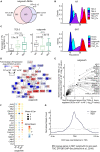
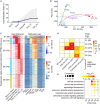
Trophoblast stem cells (TSCs) are critical to mammalian embryogenesis by providing the cell source of the placenta. TSCs can be derived from trophoblast cells. However, the efficiency of TSC derivation from somatic cell nuclear transfer (NT) blastocysts is low. The regulatory mechanisms underlying transcription dynamics and epigenetic landscape remodeling during TSC derivation remain elusive. Here, we derived TSCs from the blastocysts by natural fertilization (NF), NT, and a histone deacetylase inhibitor Scriptaid-treated NT (SNT). Profiling of the transcriptomes across the stages of TSC derivation revealed that fibroblast growth factor 4 (FGF4) treatment resulted in many differentially expressed genes (DEGs) at outgrowth and initiated transcription program for TSC formation. We identified 75 transcription factors (TFs) that are continuously upregulated during NF TSC derivation, whose transcription profiles can infer the time course of NF not NT TSC derivation. Most DEGs in NT outgrowth are rescued in SNT outgrowth. The correct time course of SNT TSC derivation is inferred accordingly. Moreover, these TFs comprise an interaction network important to TSC stemness. Profiling of DNA methylation dynamics showed an extremely low level before FGF4 treatment and gradual increases afterward. FGF4 treatment results in a distinct DNA methylation remodeling process committed to TSC formation. We further identified 1,293 CpG islands (CGIs) whose DNA methylation difference is more than 0.25 during NF TSC derivation. The majority of these CGIs become highly methylated upon FGF4 treatment and remain in high levels. This may create a barrier for lineage commitment to restrict embryonic development, and ensure TSC formation. There exist hundreds of aberrantly methylated CGIs during NT TSC derivation, most of which are corrected during SNT TSC derivation. More than half of the aberrantly methylated CGIs before NT TSC formation are inherited from the donor genome. In contrast, the aberrantly methylated CGIs upon TSC formation are mainly from the highly methylated CGIs induced by FGF4 treatment. Functional annotation indicates that the aberrantly highly methylated CGIs play a role in repressing placenta development genes, etc., related to post-implantation development and maintaining TSC pluripotency. Collectively, our findings provide novel insights into the transcription dynamics, DNA methylation remodeling, and the role of FGF4 during TSC derivation.
Keywords: DNA methylation; Scriptaid; somatic cell nuclear transfer; transcriptome; trophoblast stem cell.
Copyright © 2021 Sun, Zheng, Liu, Kou, Zhao, Liang, Wang, Zhang, Xiao, Gao, Gao and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Stem cell insights into human trophoblast lineage differentiation.Hum Reprod Update. 2016 Dec;23(1):77-103. doi: 10.1093/humupd/dmw026. Epub 2016 Sep 2. Hum Reprod Update. 2016. PMID: 27591247 Review.
-
Methylation patterns in 5' terminal regions of pluripotency-related genes in bovine in vitro fertilized and cloned embryos.J Genet Genomics. 2010 May;37(5):297-304. doi: 10.1016/S1673-8527(09)60047-3. J Genet Genomics. 2010. PMID: 20513630
-
Trophectoderm differentiation to invasive syncytiotrophoblast is promoted by endometrial epithelial cells during human embryo implantation.Hum Reprod. 2022 Apr 1;37(4):777-792. doi: 10.1093/humrep/deac008. Hum Reprod. 2022. PMID: 35079788 Free PMC article.
-
Super Enhancer Profiles Identify Key Cell Identity Genes During Differentiation From Embryonic Stem Cells to Trophoblast Stem Cells Super Enhencers in Trophoblast Differentiation.Front Genet. 2021 Oct 12;12:762529. doi: 10.3389/fgene.2021.762529. eCollection 2021. Front Genet. 2021. PMID: 34712273 Free PMC article.
-
Trophoblast stem cells.Biol Reprod. 2011 Mar;84(3):412-21. doi: 10.1095/biolreprod.110.088724. Epub 2010 Nov 24. Biol Reprod. 2011. PMID: 21106963 Free PMC article. Review.
Cited by
-
Conserved and divergent features of trophoblast stem cells.J Mol Endocrinol. 2024 Feb 19;72(4):e230131. doi: 10.1530/JME-23-0131. Print 2024 May 1. J Mol Endocrinol. 2024. PMID: 38276878 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

