Milieu matters: An in vitro wound milieu to recapitulate key features of, and probe new insights into, mixed-species bacterial biofilms
- PMID: 33912828
- PMCID: PMC8065265
- DOI: 10.1016/j.bioflm.2021.100047
Milieu matters: An in vitro wound milieu to recapitulate key features of, and probe new insights into, mixed-species bacterial biofilms
Abstract
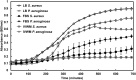
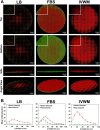
Bacterial biofilms are a major cause of delayed wound healing. Consequently, the study of wound biofilms, particularly in host-relevant conditions, has gained importance. Most in vitro studies employ refined laboratory media to study biofilms, representing conditions that are not relevant to the infection state. To mimic the wound milieu, in vitro biofilm studies often incorporate serum or plasma in growth conditions, or employ clot or matrix-based biofilm models. While incorporating serum or plasma alone is a minimalistic approach, the more complex in vitro wound models are technically demanding, and poorly compatible with standard biofilm assays. Based on previous reports of clinical wound fluid composition, we have developed an in vitro wound milieu (IVWM) that includes, in addition to serum (to recapitulate wound fluid), matrix elements and biochemical factors. With Luria-Bertani broth and Fetal Bovine Serum (FBS) for comparison, the IVWM was used to study planktonic growth, biofilm features, and interspecies interactions, of common wound pathogens, Staphylococcus aureus and Pseudomonas aeruginosa. We demonstrate that the IVWM recapitulates widely reported in vivo biofilm features such as biomass formation, metabolic activity, increased antibiotic tolerance, 3D structure, and interspecies interactions for monospecies and mixed-species biofilms. Further, the IVWM is simple to formulate, uses laboratory-grade components, and is compatible with standard biofilm assays. Given this, it holds potential as a tractable approach to study wound biofilms under host-relevant conditions.
Keywords: Biofilms; Interspecies interactions; Milieu; Mixed-species; Pseudomonas aeruginosa; Staphylococcus aureus; Wounds.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Adding a new dimension: Multi-level structure and organization of mixed-species Pseudomonas aeruginosa and Staphylococcus aureus biofilms in a 4-D wound microenvironment.Biofilm. 2022 Oct 20;4:100087. doi: 10.1016/j.bioflm.2022.100087. eCollection 2022 Dec. Biofilm. 2022. PMID: 36324526 Free PMC article.
-
Culture Shock: An Investigation into the Tolerance of Pathogenic Biofilms to Antiseptics in Environments Resembling the Chronic Wound Milieu.Int J Mol Sci. 2023 Dec 8;24(24):17242. doi: 10.3390/ijms242417242. Int J Mol Sci. 2023. PMID: 38139071 Free PMC article.
-
The chronic wound milieu changes essential oils' antibiofilm activity-an in vitro and larval model study.Sci Rep. 2024 Jan 26;14(1):2218. doi: 10.1038/s41598-024-52424-6. Sci Rep. 2024. PMID: 38278929 Free PMC article.
-
Metabolic modeling of a chronic wound biofilm consortium predicts spatial partitioning of bacterial species.BMC Syst Biol. 2016 Sep 7;10(1):90. doi: 10.1186/s12918-016-0334-8. BMC Syst Biol. 2016. PMID: 27604263 Free PMC article.
-
Matrix Polysaccharides and SiaD Diguanylate Cyclase Alter Community Structure and Competitiveness of Pseudomonas aeruginosa during Dual-Species Biofilm Development with Staphylococcus aureus.mBio. 2018 Nov 6;9(6):e00585-18. doi: 10.1128/mBio.00585-18. mBio. 2018. PMID: 30401769 Free PMC article.
Cited by
-
3D printed personalized wound dressings using a hydrophobic deep eutectic solvent (HDES)-formulated emulgel.RSC Adv. 2024 Oct 28;14(46):34175-34191. doi: 10.1039/d4ra05456c. eCollection 2024 Oct 23. RSC Adv. 2024. PMID: 39469022 Free PMC article.
-
Tools of the Trade: Image Analysis Programs for Confocal Laser-Scanning Microscopy Studies of Biofilms and Considerations for Their Use by Experimental Researchers.ACS Omega. 2023 May 26;8(23):20163-20177. doi: 10.1021/acsomega.2c07255. eCollection 2023 Jun 13. ACS Omega. 2023. PMID: 37332792 Free PMC article. Review.
-
Larval Therapy and Larval Excretions/Secretions: A Potential Treatment for Biofilm in Chronic Wounds? A Systematic Review.Microorganisms. 2023 Feb 11;11(2):457. doi: 10.3390/microorganisms11020457. Microorganisms. 2023. PMID: 36838422 Free PMC article. Review.
-
Plasma activated water as a pre-treatment strategy in the context of biofilm-infected chronic wounds.Biofilm. 2023 Sep 19;6:100154. doi: 10.1016/j.bioflm.2023.100154. eCollection 2023 Dec 15. Biofilm. 2023. PMID: 37771391 Free PMC article.
-
Adding a new dimension: Multi-level structure and organization of mixed-species Pseudomonas aeruginosa and Staphylococcus aureus biofilms in a 4-D wound microenvironment.Biofilm. 2022 Oct 20;4:100087. doi: 10.1016/j.bioflm.2022.100087. eCollection 2022 Dec. Biofilm. 2022. PMID: 36324526 Free PMC article.
References
-
- Uluer E.T., Vatansever H.S., Kurt F.Ő. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2017. Wound healing and microenvironment; pp. 67–77. Wound heal. - DOI
-
- Manuela B., Milad K., Anna-Lena S., Julian-Dario R., Ewa Klara S. Acute and chronic wound fluid inversely influence wound healing in an in-vitro 3D wound model. J Tissue Repair Regen. 2017;1:1. doi: 10.14302/issn.2640-6403.jtrr-17-1818. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

