Updated and Validated Pan-Coronavirus PCR Assay to Detect All Coronavirus Genera
- PMID: 33915875
- PMCID: PMC8067199
- DOI: 10.3390/v13040599
Updated and Validated Pan-Coronavirus PCR Assay to Detect All Coronavirus Genera
Abstract
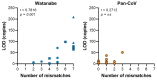
Coronavirus (CoV) spillover events from wildlife reservoirs can result in mild to severe human respiratory illness. These spillover events underlie the importance of detecting known and novel CoVs circulating in reservoir host species and determining CoV prevalence and distribution, allowing improved prediction of spillover events or where a human-reservoir interface should be closely monitored. To increase the likelihood of detecting all circulating genera and strains, we have modified primers published by Watanabe et al. in 2010 to generate a semi-nested pan-CoV PCR assay. Representatives from the four coronavirus genera (α-CoVs, β-CoVs, γ-CoVs and δ-CoVs) were tested and all of the in-house CoVs were detected using this assay. After comparing both assays, we found that the updated assay reliably detected viruses in all genera of CoVs with high sensitivity, whereas the sensitivity of the original assay was lower. Our updated PCR assay is an important tool to detect, monitor and track CoVs to enhance viral surveillance in reservoir hosts.
Keywords: PCR; coronavirus; detection; pan-CoV PCR; pandemic; reservoir host.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens.Viruses. 2024 Mar 29;16(4):534. doi: 10.3390/v16040534. Viruses. 2024. PMID: 38675878 Free PMC article.
-
Development of a one-step RT-PCR assay for detection of pancoronaviruses (α-, β-, γ-, and δ-coronaviruses) using newly designed degenerate primers for porcine and avian `fecal samples.J Virol Methods. 2018 Jun;256:116-122. doi: 10.1016/j.jviromet.2018.02.021. Epub 2018 Feb 27. J Virol Methods. 2018. PMID: 29499225 Free PMC article.
-
Epidemiology of Deltacoronaviruses (δ-CoV) and Gammacoronaviruses (γ-CoV) in Wild Birds in the United States.Viruses. 2019 Sep 26;11(10):897. doi: 10.3390/v11100897. Viruses. 2019. PMID: 31561462 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Naturally Occurring Animal Coronaviruses as Models for Studying Highly Pathogenic Human Coronaviral Disease.Vet Pathol. 2021 May;58(3):438-452. doi: 10.1177/0300985820980842. Epub 2020 Dec 28. Vet Pathol. 2021. PMID: 33357102 Review.
Cited by
-
Best Molecular Tools to Investigate Coronavirus Diversity in Mammals: A Comparison.Viruses. 2021 Oct 1;13(10):1975. doi: 10.3390/v13101975. Viruses. 2021. PMID: 34696405 Free PMC article. Review.
-
Genetic diversity among reptilian orthoreoviruses isolated from pet snakes and lizards.Front Vet Sci. 2023 Feb 2;10:1058133. doi: 10.3389/fvets.2023.1058133. eCollection 2023. Front Vet Sci. 2023. PMID: 36816198 Free PMC article.
-
Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens.Viruses. 2024 Mar 29;16(4):534. doi: 10.3390/v16040534. Viruses. 2024. PMID: 38675878 Free PMC article.
-
Survey of white-footed mice in Connecticut, USA reveals low SARS-CoV-2 seroprevalence and infection with divergent betacoronaviruses.bioRxiv [Preprint]. 2023 Sep 27:2023.09.22.559030. doi: 10.1101/2023.09.22.559030. bioRxiv. 2023. PMID: 37808797 Free PMC article. Preprint.
-
Severe acute respiratory disease in American mink experimentally infected with SARS-CoV-2.JCI Insight. 2022 Nov 22;7(22):e159573. doi: 10.1172/jci.insight.159573. JCI Insight. 2022. PMID: 36509288 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

