TbTRF suppresses the TERRA level and regulates the cell cycle-dependent TERRA foci number with a TERRA binding activity in its C-terminal Myb domain
- PMID: 34048580
- PMCID: PMC8191777
- DOI: 10.1093/nar/gkab401
TbTRF suppresses the TERRA level and regulates the cell cycle-dependent TERRA foci number with a TERRA binding activity in its C-terminal Myb domain
Abstract
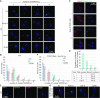
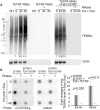
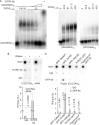
Telomere repeat-containing RNA (TERRA) has been identified in multiple organisms including Trypanosoma brucei, a protozoan parasite that causes human African trypanosomiasis. T. brucei regularly switches its major surface antigen, VSG, to evade the host immune response. VSG is expressed exclusively from subtelomeric expression sites, and we have shown that telomere proteins play important roles in the regulation of VSG silencing and switching. In this study, we identify several unique features of TERRA and telomere biology in T. brucei. First, the number of TERRA foci is cell cycle-regulated and influenced by TbTRF, the duplex telomere DNA binding factor in T. brucei. Second, TERRA is transcribed by RNA polymerase I mainly from a single telomere downstream of the active VSG. Third, TbTRF binds TERRA through its C-terminal Myb domain, which also has the duplex DNA binding activity, in a sequence-specific manner and suppresses the TERRA level without affecting its half-life. Finally, levels of the telomeric R-loop and telomere DNA damage were increased upon TbTRF depletion. Overexpression of an ectopic allele of RNase H1 that resolves the R-loop structure in TbTRF RNAi cells can partially suppress these phenotypes, revealing an underlying mechanism of how TbTRF helps maintain telomere integrity.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities.Pathogens. 2021 Jul 30;10(8):967. doi: 10.3390/pathogens10080967. Pathogens. 2021. PMID: 34451431 Free PMC article. Review.
-
Suppression of subtelomeric VSG switching by Trypanosoma brucei TRF requires its TTAGGG repeat-binding activity.Nucleic Acids Res. 2014 Nov 10;42(20):12899-911. doi: 10.1093/nar/gku942. Epub 2014 Oct 13. Nucleic Acids Res. 2014. PMID: 25313155 Free PMC article.
-
Trypanosoma brucei RAP1 maintains telomere and subtelomere integrity by suppressing TERRA and telomeric RNA:DNA hybrids.Nucleic Acids Res. 2017 Jun 2;45(10):5785-5796. doi: 10.1093/nar/gkx184. Nucleic Acids Res. 2017. PMID: 28334836 Free PMC article.
-
Trypanosoma brucei TIF2 and TRF Suppress VSG Switching Using Overlapping and Independent Mechanisms.PLoS One. 2016 Jun 3;11(6):e0156746. doi: 10.1371/journal.pone.0156746. eCollection 2016. PLoS One. 2016. PMID: 27258069 Free PMC article.
-
Telomere and Subtelomere R-loops and Antigenic Variation in Trypanosomes.J Mol Biol. 2020 Jul 10;432(15):4167-4185. doi: 10.1016/j.jmb.2019.10.025. Epub 2019 Nov 2. J Mol Biol. 2020. PMID: 31682833 Free PMC article. Review.
Cited by
-
Unwrap RAP1's Mystery at Kinetoplastid Telomeres.Biomolecules. 2024 Jan 4;14(1):67. doi: 10.3390/biom14010067. Biomolecules. 2024. PMID: 38254667 Free PMC article. Review.
-
Regulation of Antigenic Variation by Trypanosoma brucei Telomere Proteins Depends on Their Unique DNA Binding Activities.Pathogens. 2021 Jul 30;10(8):967. doi: 10.3390/pathogens10080967. Pathogens. 2021. PMID: 34451431 Free PMC article. Review.
-
POLIE suppresses telomerase-mediated telomere G-strand extension and helps ensure proper telomere C-strand synthesis in trypanosomes.Nucleic Acids Res. 2022 Feb 28;50(4):2036-2050. doi: 10.1093/nar/gkac023. Nucleic Acids Res. 2022. PMID: 35061898 Free PMC article.
-
The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.Nat Commun. 2023 Mar 22;14(1):1576. doi: 10.1038/s41467-023-37307-0. Nat Commun. 2023. PMID: 36949076 Free PMC article.
-
Telomere maintenance in African trypanosomes.Front Mol Biosci. 2023 Nov 24;10:1302557. doi: 10.3389/fmolb.2023.1302557. eCollection 2023. Front Mol Biosci. 2023. PMID: 38074093 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

