Marseilleviruses: An Update in 2021
- PMID: 34149639
- PMCID: PMC8208085
- DOI: 10.3389/fmicb.2021.648731
Marseilleviruses: An Update in 2021
Abstract
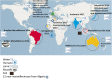
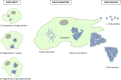
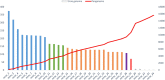
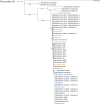
The family Marseilleviridae was the second family of giant viruses that was described in 2013, after the family Mimiviridae. Marseillevirus marseillevirus, isolated in 2007 by coculture on Acanthamoeba polyphaga, is the prototype member of this family. Afterward, the worldwide distribution of marseilleviruses was revealed through their isolation from samples of various types and sources. Thus, 62 were isolated from environmental water, one from soil, one from a dipteran, one from mussels, and two from asymptomatic humans, which led to the description of 67 marseillevirus isolates, including 21 by the IHU Méditerranée Infection in France. Recently, five marseillevirus genomes were assembled from deep sea sediment in Norway. Isolated marseilleviruses have ≈250 nm long icosahedral capsids and 348-404 kilobase long mosaic genomes that encode 386-545 predicted proteins. Comparative genomic analyses indicate that the family Marseilleviridae includes five lineages and possesses a pangenome composed of 3,082 clusters of genes. The detection of marseilleviruses in both symptomatic and asymptomatic humans in stool, blood, and lymph nodes, and an up-to-30-day persistence of marseillevirus in rats and mice, raise questions concerning their possible clinical significance that are still under investigation.
Keywords: Marseilleviridae; Pimascovirales; amoeba; giant virus; human; marseillevirus; megavirales.
Copyright © 2021 Sahmi-Bounsiar, Rolland, Aherfi, Boudjemaa, Levasseur, La Scola and Colson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The expanding family Marseilleviridae.Virology. 2014 Oct;466-467:27-37. doi: 10.1016/j.virol.2014.07.014. Epub 2014 Aug 5. Virology. 2014. PMID: 25104553 Review.
-
A Brazilian Marseillevirus Is the Founding Member of a Lineage in Family Marseilleviridae.Viruses. 2016 Mar 10;8(3):76. doi: 10.3390/v8030076. Viruses. 2016. PMID: 26978387 Free PMC article.
-
Complete genome sequence of Cannes 8 virus, a new member of the proposed family "Marseilleviridae".Virus Genes. 2013 Dec;47(3):550-5. doi: 10.1007/s11262-013-0965-4. Epub 2013 Aug 4. Virus Genes. 2013. PMID: 23912978
-
"Marseilleviridae", a new family of giant viruses infecting amoebae.Arch Virol. 2013 Apr;158(4):915-20. doi: 10.1007/s00705-012-1537-y. Epub 2012 Nov 29. Arch Virol. 2013. PMID: 23188494
-
Giant Viruses of Amoebas: An Update.Front Microbiol. 2016 Mar 22;7:349. doi: 10.3389/fmicb.2016.00349. eCollection 2016. Front Microbiol. 2016. PMID: 27047465 Free PMC article. Review.
Cited by
-
The genomic and phylogenetic analysis of Marseillevirus cajuinensis raises questions about the evolution of Marseilleviridae lineages and their taxonomical organization.J Virol. 2024 Jun 13;98(6):e0051324. doi: 10.1128/jvi.00513-24. Epub 2024 May 16. J Virol. 2024. PMID: 38752754 Free PMC article.
-
Analysis of the Genomic Features and Evolutionary History of Pithovirus-Like Isolates Reveals Two Major Divergent Groups of Viruses.J Virol. 2023 Jul 27;97(7):e0041123. doi: 10.1128/jvi.00411-23. Epub 2023 Jul 3. J Virol. 2023. PMID: 37395647 Free PMC article.
-
Giant Viruses as a Source of Novel Enzymes for Biotechnological Application.Pathogens. 2022 Dec 1;11(12):1453. doi: 10.3390/pathogens11121453. Pathogens. 2022. PMID: 36558786 Free PMC article. Review.
-
Amoebal Tubulin Cleavage Late during Infection Is a Characteristic Feature of Mimivirus but Not of Marseillevirus.Microbiol Spectr. 2022 Dec 21;10(6):e0275322. doi: 10.1128/spectrum.02753-22. Epub 2022 Dec 1. Microbiol Spectr. 2022. PMID: 36453900 Free PMC article.
-
Elimination of Foreign Sequences in Eukaryotic Viral Reference Genomes Improves the Accuracy of Virome Analysis.mSystems. 2022 Dec 20;7(6):e0090722. doi: 10.1128/msystems.00907-22. Epub 2022 Oct 26. mSystems. 2022. PMID: 36286492 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

