Protein lysine crotonylation: past, present, perspective
- PMID: 34262024
- PMCID: PMC8280118
- DOI: 10.1038/s41419-021-03987-z
Protein lysine crotonylation: past, present, perspective
Abstract
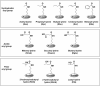
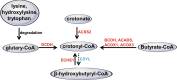
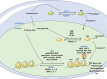
Lysine crotonylation has been discovered in histone and non-histone proteins and found to be involved in diverse diseases and biological processes, such as neuropsychiatric disease, carcinogenesis, spermatogenesis, tissue injury, and inflammation. The unique carbon-carbon π-bond structure indicates that lysine crotonylation may use distinct regulatory mechanisms from the widely studied other types of lysine acylation. In this review, we discussed the regulation of lysine crotonylation by enzymatic and non-enzymatic mechanisms, the recognition of substrate proteins, the physiological functions of lysine crotonylation and its cross-talk with other types of modification. The tools and methods for prediction and detection of lysine crotonylation were also described.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Functions and mechanisms of lysine crotonylation.J Cell Mol Med. 2019 Nov;23(11):7163-7169. doi: 10.1111/jcmm.14650. Epub 2019 Sep 1. J Cell Mol Med. 2019. PMID: 31475443 Free PMC article. Review.
-
The Function and related Diseases of Protein Crotonylation.Int J Biol Sci. 2021 Aug 9;17(13):3441-3455. doi: 10.7150/ijbs.58872. eCollection 2021. Int J Biol Sci. 2021. PMID: 34512158 Free PMC article. Review.
-
Assays for Acetylation and Other Acylations of Lysine Residues.Curr Protoc Protein Sci. 2017 Feb 2;87:14.11.1-14.11.18. doi: 10.1002/cpps.26. Curr Protoc Protein Sci. 2017. PMID: 28150880
-
Large-scale lysine crotonylation analysis reveals its potential role in spermiogenesis in the Chinese mitten crab Eriocheir sinensis.J Proteomics. 2020 Aug 30;226:103891. doi: 10.1016/j.jprot.2020.103891. Epub 2020 Jul 4. J Proteomics. 2020. PMID: 32629196
-
Beyond histone acetylation-writing and erasing histone acylations.Curr Opin Struct Biol. 2018 Dec;53:169-177. doi: 10.1016/j.sbi.2018.10.001. Epub 2018 Nov 2. Curr Opin Struct Biol. 2018. PMID: 30391813 Review.
Cited by
-
Current computational tools for protein lysine acylation site prediction.Brief Bioinform. 2024 Sep 23;25(6):bbae469. doi: 10.1093/bib/bbae469. Brief Bioinform. 2024. PMID: 39316944
-
Integrative transcriptome analysis identifies a crotonylation gene signature for predicting prognosis and drug sensitivity in hepatocellular carcinoma.J Cell Mol Med. 2024 Oct;28(20):e70083. doi: 10.1111/jcmm.70083. J Cell Mol Med. 2024. PMID: 39428564 Free PMC article.
-
Global crotonylome reveals hypoxia-mediated lamin A crotonylation regulated by HDAC6 in liver cancer.Cell Death Dis. 2022 Aug 17;13(8):717. doi: 10.1038/s41419-022-05165-1. Cell Death Dis. 2022. PMID: 35977926 Free PMC article.
-
Protein modification by short-chain fatty acid metabolites in sepsis: a comprehensive review.Front Immunol. 2023 Oct 6;14:1171834. doi: 10.3389/fimmu.2023.1171834. eCollection 2023. Front Immunol. 2023. PMID: 37869005 Free PMC article. Review.
-
Regulatory Mechanism of Protein Crotonylation and Its Relationship with Cancer.Cells. 2024 Nov 2;13(21):1812. doi: 10.3390/cells13211812. Cells. 2024. PMID: 39513918 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

