Approaches to Enhance Precise CRISPR/Cas9-Mediated Genome Editing
- PMID: 34445274
- PMCID: PMC8395304
- DOI: 10.3390/ijms22168571
Approaches to Enhance Precise CRISPR/Cas9-Mediated Genome Editing
Abstract
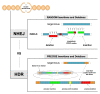
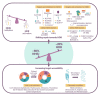
Modification of the human genome has immense potential for preventing or treating disease. Modern genome editing techniques based on CRISPR/Cas9 show great promise for altering disease-relevant genes. The efficacy of precision editing at CRISPR/Cas9-induced double-strand breaks is dependent on the relative activities of nuclear DNA repair pathways, including the homology-directed repair and error-prone non-homologous end-joining pathways. The competition between multiple DNA repair pathways generates mosaic and/or therapeutically undesirable editing outcomes. Importantly, genetic models have validated key DNA repair pathways as druggable _targets for increasing editing efficacy. In this review, we highlight approaches that can be used to achieve the desired genome modification, including the latest progress using small molecule modulators and engineered CRISPR/Cas proteins to enhance precision editing.
Keywords: CRISPR/Cas9; engineered Cas9; genome editing; homology-directed repair; small molecules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Precision genome editing in the CRISPR era.Biochem Cell Biol. 2017 Apr;95(2):187-201. doi: 10.1139/bcb-2016-0137. Epub 2016 Sep 29. Biochem Cell Biol. 2017. PMID: 28177771 Review.
-
Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing.Nat Biomed Eng. 2017 Nov;1(11):878-888. doi: 10.1038/s41551-017-0145-2. Epub 2017 Oct 9. Nat Biomed Eng. 2017. PMID: 31015609 Free PMC article.
-
Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks.Int J Mol Sci. 2020 Sep 4;21(18):6461. doi: 10.3390/ijms21186461. Int J Mol Sci. 2020. PMID: 32899704 Free PMC article. Review.
-
Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing.Genome Biol. 2018 Oct 19;19(1):170. doi: 10.1186/s13059-018-1518-x. Genome Biol. 2018. PMID: 30340517 Free PMC article.
-
Proximal binding of dCas9 at a DNA double strand break stimulates homology-directed repair as a local inhibitor of classical non-homologous end joining.Nucleic Acids Res. 2023 Apr 11;51(6):2740-2758. doi: 10.1093/nar/gkad116. Nucleic Acids Res. 2023. PMID: 36864759 Free PMC article.
Cited by
-
Genetically modified mice as a tool for the study of human diseases.Mol Biol Rep. 2024 Jan 18;51(1):135. doi: 10.1007/s11033-023-09066-0. Mol Biol Rep. 2024. PMID: 38236499 Review.
-
CRISPR Ribonucleoprotein-Mediated Precise Editing of Multiple Genes in Porcine Fibroblasts.Animals (Basel). 2024 Feb 18;14(4):650. doi: 10.3390/ani14040650. Animals (Basel). 2024. PMID: 38396618 Free PMC article.
-
Zebrafish CCNF and FUS Mediate Stress-Specific Motor Responses.Cells. 2024 Feb 21;13(5):372. doi: 10.3390/cells13050372. Cells. 2024. PMID: 38474336 Free PMC article.
-
CRISPR/Cas as a Genome-Editing Technique in Fruit Tree Breeding.Int J Mol Sci. 2023 Nov 23;24(23):16656. doi: 10.3390/ijms242316656. Int J Mol Sci. 2023. PMID: 38068981 Free PMC article. Review.
-
Efficient delivery of a large-size Cas9-EGFP vector in porcine fetal fibroblasts using a Lonza 4D-Nucleofector system.BMC Biotechnol. 2023 Aug 16;23(1):29. doi: 10.1186/s12896-023-00799-1. BMC Biotechnol. 2023. PMID: 37587435 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

