Best Molecular Tools to Investigate Coronavirus Diversity in Mammals: A Comparison
- PMID: 34696405
- PMCID: PMC8538982
- DOI: 10.3390/v13101975
Best Molecular Tools to Investigate Coronavirus Diversity in Mammals: A Comparison
Abstract
Coronaviruses (CoVs) are widespread and highly diversified in wildlife and domestic mammals and can emerge as zoonotic or epizootic pathogens and consequently host shift from these reservoirs, highlighting the importance of veterinary surveillance. All genera can be found in mammals, with α and β showing the highest frequency and diversification. The aims of this study were to review the literature for features of CoV surveillance in animals, to test widely used molecular protocols, and to identify the most effective one in terms of spectrum and sensitivity. We combined a literature review with analyses in silico and in vitro using viral strains and archive field samples. We found that most protocols defined as pan-coronavirus are strongly biased towards α- and β-CoVs and show medium-low sensitivity. The best results were observed using our new protocol, showing LoD 100 PFU/mL for SARS-CoV-2, 50 TCID50/mL for CaCoV, 0.39 TCID50/mL for BoCoV, and 9 ± 1 log2 ×10-5 HA for IBV. The protocol successfully confirmed the positivity for a broad range of CoVs in 30/30 field samples. Our study points out that pan-CoV surveillance in mammals could be strongly improved in sensitivity and spectrum and propose the application of a new RT-PCR assay, which is able to detect CoVs from all four genera, with an optimal sensitivity for α-, β-, and γ-.
Keywords: BoCoV; CaCoV; RT-PCR; SARS-CoV-2; coronavirus; pan-CoV; surveillance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
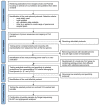

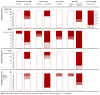

Similar articles
-
Full Genome of batCoV/MinFul/2018/SriLanka, a Novel Alpha-Coronavirus Detected in Miniopterus fuliginosus, Sri Lanka.Viruses. 2022 Feb 7;14(2):337. doi: 10.3390/v14020337. Viruses. 2022. PMID: 35215931 Free PMC article.
-
Coronavirus surveillance of wildlife in the Lao People's Democratic Republic detects viral RNA in rodents.Arch Virol. 2020 Aug;165(8):1869-1875. doi: 10.1007/s00705-020-04683-7. Epub 2020 Jun 1. Arch Virol. 2020. PMID: 32488616 Free PMC article.
-
Longitudinal monitoring in Cambodia suggests higher circulation of alpha and betacoronaviruses in juvenile and immature bats of three species.Sci Rep. 2021 Dec 17;11(1):24145. doi: 10.1038/s41598-021-03169-z. Sci Rep. 2021. PMID: 34921180 Free PMC article.
-
Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: A case study of bats.PLoS Pathog. 2020 Sep 3;16(9):e1008758. doi: 10.1371/journal.ppat.1008758. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32881980 Free PMC article. Review.
-
Global Epidemiology of Bat Coronaviruses.Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. Viruses. 2019. PMID: 30791586 Free PMC article. Review.
Cited by
-
Canine Distemper Virus in Sardinia, Italy: Detection and Phylogenetic Analysis in Foxes.Animals (Basel). 2024 Oct 31;14(21):3134. doi: 10.3390/ani14213134. Animals (Basel). 2024. PMID: 39518857 Free PMC article.
-
Molecular and Serological Detection of Bovine Coronaviruses in Marmots (Marmota marmota) in the Alpine Region.Viruses. 2024 Apr 11;16(4):591. doi: 10.3390/v16040591. Viruses. 2024. PMID: 38675932 Free PMC article.
-
Molecular Detection and Characterization of Coronaviruses in Migratory Ducks from Portugal Show the Circulation of Gammacoronavirus and Deltacoronavirus.Animals (Basel). 2022 Nov 25;12(23):3283. doi: 10.3390/ani12233283. Animals (Basel). 2022. PMID: 36496804 Free PMC article.
-
First Report of Alphacoronavirus Circulating in Cavernicolous Bats from Portugal.Viruses. 2023 Jul 8;15(7):1521. doi: 10.3390/v15071521. Viruses. 2023. PMID: 37515207 Free PMC article.
-
Comparative Performance in the Detection of Four Coronavirus Genera from Human, Animal, and Environmental Specimens.Viruses. 2024 Mar 29;16(4):534. doi: 10.3390/v16040534. Viruses. 2024. PMID: 38675878 Free PMC article.
References
-
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Wang M., Lam C.S.F., Xu H., Guo R., Chan K., Zheng B., et al. Complete Genome Sequence of Bat Coronavirus HKU2 from Chinese Horseshoe Bats Revealed a Much Smaller Spike Gene with a Different Evolutionary Lineage from the Rest of the Genome. Virology. 2007;367:428–439. doi: 10.1016/j.virol.2007.06.009. - DOI - PMC - PubMed
-
- Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., Groot R.J.D.E. Feline Coronavirus Type II Strains 79-1683 and 79-1146 Originate from a Double Recombination between Feline Coronavirus Type I and Canine Coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/JVI.72.5.4508-4514.1998. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

