Bioassay-Guided Interpretation of Antimicrobial Compounds in Kumu, a TCM Preparation From Picrasma quassioides' Stem via UHPLC-Orbitrap-Ion Trap Mass Spectrometry Combined With Fragmentation and Retention Time Calculation
- PMID: 34776978
- PMCID: PMC8581800
- DOI: 10.3389/fphar.2021.761751
Bioassay-Guided Interpretation of Antimicrobial Compounds in Kumu, a TCM Preparation From Picrasma quassioides' Stem via UHPLC-Orbitrap-Ion Trap Mass Spectrometry Combined With Fragmentation and Retention Time Calculation
Erratum in
-
Corrigendum: Bioassay-Guided Interpretation of Antimicrobial Compounds in Kumu, a TCM Preparation From Picrasma quassioides' Stem via UHPLC-Orbitrap-Ion Trap Mass Spectrometer Combined With Fragmentation and Retention Time Calculation.Front Pharmacol. 2022 Apr 25;13:897243. doi: 10.3389/fphar.2022.897243. eCollection 2022. Front Pharmacol. 2022. PMID: 35548352 Free PMC article.
Abstract
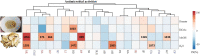
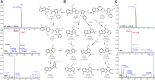
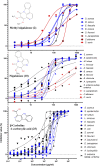
The stem of Picrasma quassioides (PQ) was recorded as a prominent traditional Chinese medicine, Kumu, which was effective for microbial infection, inflammation, fever, and dysentery, etc. At present, Kumu is widely used in China to develop different medicines, even as injection (Kumu zhusheye), for combating infections. However, the chemical basis of its antimicrobial activity has still not been elucidated. To examine the active chemicals, its stem was extracted to perform bioassay-guided purification against Staphylococcus aureus and Escherichia coli. In this study, two types of columns (normal and reverse-phase) were used for speedy bioassay-guided isolation from Kumu, and the active peaks were collected and identified via an UHPLC-Orbitrap-Ion Trap Mass Spectrometer, combined with MS Fragmenter and ChromGenius. For identification, the COCONUT Database (largest database of natural products) and a manually built PQ database were used, in combination with prediction and calculation of mass fragmentation and retention time to better infer their structures, especially for isomers. Moreover, three standards were analyzed under different conditions for developing and validating the MS method. A total of 25 active compounds were identified, including 24 alkaloids and 1 triterpenoid against S. aureus, whereas only β-carboline-1-carboxylic acid and picrasidine S were active against E. coli. Here, the good antimicrobial activity of 18 chemicals was reported for the first time. Furthermore, the spectrum of three abundant β-carbolines was assessed via their IC50 and MBC against various human pathogens. All of them exhibited strong antimicrobial activities with good potential to be developed as antibiotics. This study clearly showed the antimicrobial chemical basis of Kumu, and the results demonstrated that HRMS coupled with MS Fragmenter and ChromGenius was a powerful tool for compound analysis, which can be used for other complex samples. Beta-carbolines reported here are important lead compounds in antibiotic discovery.
Keywords: MS Fragmenter; Picrasma quassioides; beta-carboline; fragmentation prediction; kumu; orbitrap elite.
Copyright © 2021 Hu, Hu, Peng, Ghosh, Khan, Sun and Luyten.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Corrigendum: Bioassay-Guided Interpretation of Antimicrobial Compounds in Kumu, a TCM Preparation From Picrasma quassioides' Stem via UHPLC-Orbitrap-Ion Trap Mass Spectrometer Combined With Fragmentation and Retention Time Calculation.Front Pharmacol. 2022 Apr 25;13:897243. doi: 10.3389/fphar.2022.897243. eCollection 2022. Front Pharmacol. 2022. PMID: 35548352 Free PMC article.
-
Picrasma quassioides leaves: Insights from chemical profiling and bioactivity comparison with stems.Fitoterapia. 2024 Sep;177:106108. doi: 10.1016/j.fitote.2024.106108. Epub 2024 Jul 2. Fitoterapia. 2024. PMID: 38964561
-
Quality Assessment of Kumu Injection, a Traditional Chinese Medicine Preparation, Using HPLC Combined with Chemometric Methods and Qualitative and Quantitative Analysis of Multiple Alkaloids by Single Marker.Molecules. 2018 Apr 9;23(4):856. doi: 10.3390/molecules23040856. Molecules. 2018. PMID: 29642544 Free PMC article.
-
Phytochemistry, Traditional Use and Pharmacological Activity of Picrasma quassioides: A Critical Reviews.Nutrients. 2020 Aug 26;12(9):2584. doi: 10.3390/nu12092584. Nutrients. 2020. PMID: 32858812 Free PMC article. Review.
-
Pharmacological effects of Picrasma quassioides (D. Don) Benn for inflammation, cancer and neuroprotection (Review).Exp Ther Med. 2021 Dec;22(6):1357. doi: 10.3892/etm.2021.10792. Epub 2021 Sep 24. Exp Ther Med. 2021. PMID: 34659503 Free PMC article. Review.
Cited by
-
Metabolic profiling of Chimonanthus grammatus via UHPLC-HRMS-MS with computer-assisted structure elucidation and its antimicrobial activity.Front Plant Sci. 2023 May 9;14:1138913. doi: 10.3389/fpls.2023.1138913. eCollection 2023. Front Plant Sci. 2023. PMID: 37229132 Free PMC article.
-
Ethnobotanical study of Hakka traditional medicine in Ganzhou, China and their antibacterial, antifungal, and cytotoxic assessments.BMC Complement Med Ther. 2022 Sep 19;22(1):244. doi: 10.1186/s12906-022-03712-z. BMC Complement Med Ther. 2022. PMID: 36123737 Free PMC article.
-
Exploration of Type III effector Xanthomonas outer protein Q (XopQ) inhibitor from Picrasma quassioides as an antibacterial agent using chemoinformatics analysis.PLoS One. 2024 Jun 18;19(6):e0302105. doi: 10.1371/journal.pone.0302105. eCollection 2024. PLoS One. 2024. PMID: 38889115 Free PMC article.
-
Pharmacokinetic Study of Four Major Bioactive Components of Liandan Xiaoyan Formula in Ulcerative Colitis and Control Rats Using UPLC-MS/MS.Front Pharmacol. 2022 Jul 4;13:936846. doi: 10.3389/fphar.2022.936846. eCollection 2022. Front Pharmacol. 2022. PMID: 35860031 Free PMC article.
References
-
- Allen F., Greiner R., Wishart D. (2014). Competitive Fragmentation Modeling of ESI-MS/MS Spectra for Putative Metabolite Identification. Metabolomics 11, 98–110. 10.1007/s11306-014-0676-4 - DOI
-
- Amanulla S. S. D., Veerakumar S., Ramanathan K. (2017). Screening of Potential Plant Compounds as Survivin Inhibitors and its Anti-Cancer Efficacy by Molecular Docking. Cei 13, 41–48. 10.2174/1573408012666160727165940 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

