Effect of short-term hindlimb immobilization on skeletal muscle atrophy and the transcriptome in a low compared with high responder to endurance training model
- PMID: 35025912
- PMCID: PMC8757917
- DOI: 10.1371/journal.pone.0261723
Effect of short-term hindlimb immobilization on skeletal muscle atrophy and the transcriptome in a low compared with high responder to endurance training model
Abstract
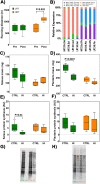
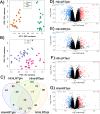
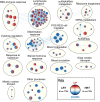
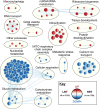
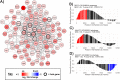
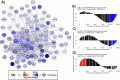
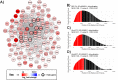
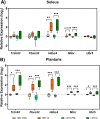
Skeletal muscle atrophy is a physiological response to disuse, aging, and disease. We compared changes in muscle mass and the transcriptome profile after short-term immobilization in a divergent model of high and low responders to endurance training to identify biological processes associated with the early atrophy response. Female rats selectively bred for high response to endurance training (HRT) and low response to endurance training (LRT; n = 6/group; generation 19) underwent 3 day hindlimb cast immobilization to compare atrophy of plantaris and soleus muscles with line-matched controls (n = 6/group). RNA sequencing was utilized to identify Gene Ontology Biological Processes with differential gene set enrichment. Aerobic training performed prior to the intervention showed HRT improved running distance (+60.6 ± 29.6%), while LRT were unchanged (-0.3 ± 13.3%). Soleus atrophy was greater in LRT vs. HRT (-9.0 ±8.8 vs. 6.2 ±8.2%; P<0.05) and there was a similar trend in plantaris (-16.4 ±5.6% vs. -8.5 ±7.4%; P = 0.064). A total of 140 and 118 biological processes were differentially enriched in plantaris and soleus muscles, respectively. Soleus muscle exhibited divergent LRT and HRT responses in processes including autophagy and immune response. In plantaris, processes associated with protein ubiquitination, as well as the atrogenes (Trim63 and Fbxo32), were more positively enriched in LRT. Overall, LRT demonstrate exacerbated atrophy compared to HRT, associated with differential gene enrichments of biological processes. This indicates that genetic factors that result in divergent adaptations to endurance exercise, may also regulate biological processes associated with short-term muscle unloading.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Low responders to endurance training exhibit impaired hypertrophy and divergent biological process responses in rat skeletal muscle.Exp Physiol. 2021 Mar;106(3):714-725. doi: 10.1113/EP089301. Epub 2021 Feb 5. Exp Physiol. 2021. PMID: 33486778 Free PMC article.
-
Physiological adaptations to resistance training in rats selectively bred for low and high response to aerobic exercise training.Exp Physiol. 2018 Nov;103(11):1513-1523. doi: 10.1113/EP087144. Epub 2018 Oct 9. Exp Physiol. 2018. PMID: 30184287 Free PMC article.
-
Running training experience attenuates disuse atrophy in fast-twitch skeletal muscles of rats.J Appl Physiol (1985). 2017 Oct 1;123(4):902-913. doi: 10.1152/japplphysiol.00289.2017. Epub 2017 Aug 3. J Appl Physiol (1985). 2017. PMID: 28775067
-
Exercise Preconditioning Blunts Early Atrogenes Expression and Atrophy in Gastrocnemius Muscle of Hindlimb Unloaded Mice.Int J Mol Sci. 2021 Dec 23;23(1):148. doi: 10.3390/ijms23010148. Int J Mol Sci. 2021. PMID: 35008572 Free PMC article.
-
Striated muscle-specific serine/threonine-protein kinase beta segregates with high versus low responsiveness to endurance exercise training.Physiol Genomics. 2020 Jan 1;52(1):35-46. doi: 10.1152/physiolgenomics.00103.2019. Epub 2019 Dec 2. Physiol Genomics. 2020. PMID: 31790338 Free PMC article.
Cited by
-
The muscle proteome reflects changes in mitochondrial function, cellular stress and proteolysis after 14 days of unilateral lower limb immobilization in active young men.PLoS One. 2022 Sep 1;17(9):e0273925. doi: 10.1371/journal.pone.0273925. eCollection 2022. PLoS One. 2022. PMID: 36048851 Free PMC article.
-
Identifying the potential therapeutic effects of miR‑6516 on muscle disuse atrophy.Mol Med Rep. 2024 Jul;30(1):119. doi: 10.3892/mmr.2024.13243. Epub 2024 May 17. Mol Med Rep. 2024. PMID: 38757344 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

