Hepatitis C Virus-Induced ROS/JNK Signaling Pathway Activates the E3 Ubiquitin Ligase Itch to Promote the Release of HCV Particles via Polyubiquitylation of VPS4A
- PMID: 35044214
- PMCID: PMC8941892
- DOI: 10.1128/JVI.01811-21
Hepatitis C Virus-Induced ROS/JNK Signaling Pathway Activates the E3 Ubiquitin Ligase Itch to Promote the Release of HCV Particles via Polyubiquitylation of VPS4A
Abstract
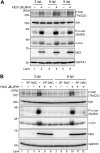
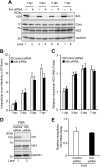
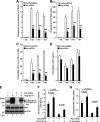
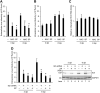
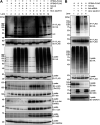
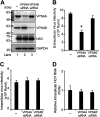
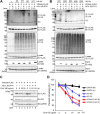
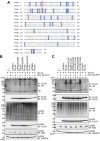
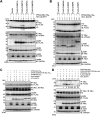
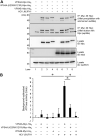
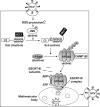
We previously reported that hepatitis C virus (HCV) infection activates the reactive oxygen species (ROS)/c-Jun N-terminal kinase (JNK) signaling pathway. However, the roles of ROS/JNK activation in the HCV life cycle remain unclear. We sought to identify a novel role of the ROS/JNK signaling pathway in the HCV life cycle. Immunoblot analysis revealed that HCV-induced ROS/JNK activation promoted phosphorylation of Itch, a HECT-type E3 ubiquitin ligase, leading to activation of Itch. The small interfering RNA (siRNA) knockdown of Itch significantly reduced the extracellular HCV infectivity titers, HCV RNA, and HCV core protein without affecting intracellular HCV infectivity titers, HCV RNA, and HCV proteins, suggesting that Itch is involved in the release of HCV particles. HCV-mediated JNK/Itch activation specifically promoted polyubiquitylation of an AAA-type ATPase, VPS4A, but not VPS4B, required to form multivesicular bodies. Site-directed mutagenesis revealed that two lysine residues (K23 and K121) on VPS4A were important for VPS4A polyubiquitylation. The siRNA knockdown of VPS4A, but not VPS4B, significantly reduced extracellular HCV infectivity titers. Coimmunoprecipitation analysis revealed that HCV infection specifically enhanced the interaction between CHMP1B, a subunit of endosomal sorting complexes required for transport (ESCRT)-III complex, and VPS4A, but not VPS4B, whereas VPS4A K23R/K121R greatly reduced the interaction with CHMP1B. HCV infection significantly increased ATPase activity of VPS4A, but not VPS4A K23R/K121R or VPS4B, suggesting that HCV-mediated polyubiquitylation of VPS4A contributes to activation of VPS4A. Taken together, we propose that the HCV-induced ROS/JNK/Itch signaling pathway promotes VPS4A polyubiquitylation, leading to enhanced VPS4A-CHMP1B interaction and promotion of VPS4A ATPase activity, thereby promoting the release of HCV particles. IMPORTANCE The ROS/JNK signaling pathway contributes to liver diseases, including steatosis, metabolic disorders, and hepatocellular carcinoma. We previously reported that HCV activates the ROS/JNK signaling pathway, leading to the enhancement of hepatic gluconeogenesis and apoptosis induction. This study further demonstrates that the HCV-induced ROS/JNK signaling pathway activates the E3 ubiquitin ligase Itch to promote release of HCV particles via polyubiquitylation of VPS4A. We provide evidence suggesting that HCV infection promotes the ROS/JNK/Itch signaling pathway and ESCRT/VPS4A machinery to release infectious HCV particles. Our results may lead to a better understanding of the mechanistic details of HCV particle release.
Keywords: ESCRT; Itch; ROS/JNK; VPS4A; hepatitis C virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Cellular Release of Infectious Hepatitis C Virus Particles via Endosomal Pathways.Viruses. 2023 Dec 14;15(12):2430. doi: 10.3390/v15122430. Viruses. 2023. PMID: 38140670 Free PMC article. Review.
-
Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition.J Biol Chem. 2010 Nov 12;285(46):35428-38. doi: 10.1074/jbc.M110.126318. Epub 2010 Aug 30. J Biol Chem. 2010. PMID: 20805225 Free PMC article.
-
Influenza virus budding does not require a functional AAA+ ATPase, VPS4.Virus Res. 2010 Oct;153(1):58-63. doi: 10.1016/j.virusres.2010.07.006. Epub 2010 Jul 17. Virus Res. 2010. PMID: 20621136 Free PMC article.
-
Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles.J Gen Virol. 2010 Feb;91(Pt 2):362-72. doi: 10.1099/vir.0.017285-0. Epub 2009 Oct 14. J Gen Virol. 2010. PMID: 19828764 Free PMC article.
-
The regulation of Endosomal Sorting Complex Required for Transport and accessory proteins in multivesicular body sorting and enveloped viral budding - An overview.Int J Biol Macromol. 2019 Apr 15;127:1-11. doi: 10.1016/j.ijbiomac.2019.01.015. Epub 2019 Jan 4. Int J Biol Macromol. 2019. PMID: 30615963 Review.
Cited by
-
Transcription Factor JunB Suppresses Hepatitis C Virus Replication.Kobe J Med Sci. 2023 Aug 31;69(3):E86-E95. Kobe J Med Sci. 2023. PMID: 37661632 Free PMC article.
-
The Early Secretory Pathway Is Crucial for Multiple Aspects of the Hepatitis C Virus Life Cycle.J Virol. 2023 Jul 27;97(7):e0018023. doi: 10.1128/jvi.00180-23. Epub 2023 Jun 20. J Virol. 2023. PMID: 37338368 Free PMC article.
-
Cellular Release of Infectious Hepatitis C Virus Particles via Endosomal Pathways.Viruses. 2023 Dec 14;15(12):2430. doi: 10.3390/v15122430. Viruses. 2023. PMID: 38140670 Free PMC article. Review.
-
Insights into the function of ESCRT and its role in enveloped virus infection.Front Microbiol. 2023 Oct 6;14:1261651. doi: 10.3389/fmicb.2023.1261651. eCollection 2023. Front Microbiol. 2023. PMID: 37869652 Free PMC article. Review.
-
Metabolomics combined with network pharmacology reveals a role for astragaloside IV in inhibiting enterovirus 71 replication via PI3K-AKT signaling.J Transl Med. 2024 Jun 10;22(1):555. doi: 10.1186/s12967-024-05355-9. J Transl Med. 2024. PMID: 38858642 Free PMC article.
References
-
- World Health Organization. 2017. Global hepatitis report. World Health Organization, Geneva, Switzerland.
-
- Mankouri J, Walter C, Stewart H, Bentham M, Park WS, Heo WD, Fukuda M, Griffin S, Harris M. 2016. Release of infectious hepatitis C virus from Huh7 cells occurs via a trans-Golgi network-to-endosome pathway independent of very-low-density lipoprotein secretion. J Virol 90:7159–7170. 10.1128/JVI.00826-16. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

