Tetramic Acids Mutanocyclin and Reutericyclin A, Produced by Streptococcus mutans Strain B04Sm5 Modulate the Ecology of an in vitro Oral Biofilm
- PMID: 35048077
- PMCID: PMC8757879
- DOI: 10.3389/froh.2021.796140
Tetramic Acids Mutanocyclin and Reutericyclin A, Produced by Streptococcus mutans Strain B04Sm5 Modulate the Ecology of an in vitro Oral Biofilm
Abstract
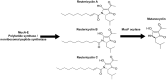
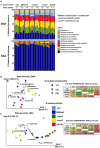
The human oral microbiome consists of diverse microbes actively communicating and interacting through a variety of biochemical mechanisms. Dental caries is a major public health issue caused by fermentable carbohydrate consumption that leads to dysbiosis of the oral microbiome. Streptococcus mutans is a known major contributor to caries pathogenesis, due to its exceptional ability to form biofilms in the presence of sucrose, as well as to its acidophilic lifestyle. S. mutans can also kill competing bacteria, which are typically health associated, through the production of bacteriocins and other small molecules. A subset of S. mutans strains encode the muc biosynthetic gene cluster (BGC), which was recently shown to produce the tetramic acids, mutanocyclin and reutericyclins A, B, and C. Reutericyclin A displayed strong antimicrobial activity and mutanocyclin appeared to be anti-inflammatory; however the effect of these compounds, and the carriage of muc by S. mutans, on the ecology of the oral microbiota is not known, and was examined here using a previously developed in vitro biofilm model derived from human saliva. While reutericyclin significantly inhibited in vitro biofilm formation and acid production at sub-nanomolar concentrations, mutanocyclin did not present any activity until the high micromolar range. 16S rRNA gene sequencing revealed that reutericyclin drastically altered the biofilm community composition, while mutanocyclin showed a more specific effect, reducing the relative abundance of cariogenic Limosilactobacillus fermentum. Mutanocyclin or reutericyclin produced by the S. mutans strains amended to the community did not appear to affect the community in the same way as the purified compounds, although the results were somewhat confounded by the differing growth rates of the S. mutans strains. Regardless of the strain added, the addition of S. mutans to the in vitro community significantly increased the abundance of S. mutans and Veillonella infantium, only. Overall, this study illustrates that reutericyclin A and mutanocyclin do impact the ecology of a complex in vitro oral biofilm; however, further research is needed to determine the extent to which the production of these compounds affects the virulence of S. mutans.
Keywords: Streptococcus mutans (S. mutans); biofilm; dental caries; oral microbiome; oral microbiota; tetramic acid.
Copyright © 2022 Uranga, Nelson, Edlund and Baker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
[Latest Findings on Polyketides/Non-ribosomal Peptides That Are Secondary Metabolites of Streptococcus mutans].Sichuan Da Xue Xue Bao Yi Xue Ban. 2023 May;54(3):685-691. doi: 10.12182/20230560302. Sichuan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37248606 Free PMC article. Review. Chinese.
-
mucG, mucH, and mucI Modulate Production of Mutanocyclin and Reutericyclins in Streptococcus mutans B04Sm5.J Bacteriol. 2022 May 17;204(5):e0004222. doi: 10.1128/jb.00042-22. Epub 2022 Apr 11. J Bacteriol. 2022. PMID: 35404110 Free PMC article.
-
Cariogenic Streptococcus mutans Produces Tetramic Acid Strain-Specific Antibiotics That Impair Commensal Colonization.ACS Infect Dis. 2020 Apr 10;6(4):563-571. doi: 10.1021/acsinfecdis.9b00365. Epub 2020 Jan 10. ACS Infect Dis. 2020. PMID: 31906623 Free PMC article.
-
Composite Long- and Short-Read Sequencing Delivers a Complete Genome Sequence of B04Sm5, a Reutericyclin- and Mutanocyclin-Producing Strain of Streptococcus mutans.Microbiol Resour Announc. 2020 Nov 19;9(47):e01067-20. doi: 10.1128/MRA.01067-20. Microbiol Resour Announc. 2020. PMID: 33214302 Free PMC article.
-
Polyketides/nonribosomal peptides from Streptococcus mutans and their ecological roles in dental biofilm.Mol Oral Microbiol. 2024 Oct;39(5):261-269. doi: 10.1111/omi.12451. Epub 2024 Jan 11. Mol Oral Microbiol. 2024. PMID: 38212261 Review.
Cited by
-
Exploring ex vivo biofilm dynamics: consequences of low ampicillin concentrations on the human oral microbiome.NPJ Biofilms Microbiomes. 2024 Apr 2;10(1):37. doi: 10.1038/s41522-024-00507-7. NPJ Biofilms Microbiomes. 2024. PMID: 38565843 Free PMC article.
-
[Latest Findings on Polyketides/Non-ribosomal Peptides That Are Secondary Metabolites of Streptococcus mutans].Sichuan Da Xue Xue Bao Yi Xue Ban. 2023 May;54(3):685-691. doi: 10.12182/20230560302. Sichuan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37248606 Free PMC article. Review. Chinese.
-
mucG, mucH, and mucI Modulate Production of Mutanocyclin and Reutericyclins in Streptococcus mutans B04Sm5.J Bacteriol. 2022 May 17;204(5):e0004222. doi: 10.1128/jb.00042-22. Epub 2022 Apr 11. J Bacteriol. 2022. PMID: 35404110 Free PMC article.
-
Commensal production of a broad-spectrum and short-lived antimicrobial peptide polyene eliminates nasal Staphylococcus aureus.Nat Microbiol. 2024 Jan;9(1):200-213. doi: 10.1038/s41564-023-01544-2. Epub 2023 Dec 18. Nat Microbiol. 2024. PMID: 38110697 Free PMC article.
-
Ecological influence by colonization of fluoride-resistant Streptococcus mutans in oral biofilm.Front Cell Infect Microbiol. 2023 Jan 9;12:1106392. doi: 10.3389/fcimb.2022.1106392. eCollection 2022. Front Cell Infect Microbiol. 2023. PMID: 36699726 Free PMC article.
References
-
- Aleti GA, Baker JL, Tang X, Alvarez R, Dinis M, Tran NC, et al. . Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. mBio. (2019) 10:e00321–e00319. 10.1128/mBio.00321-19 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

