Temporal Gene Expression Profiles Reflect the Dynamics of Lymphoid Differentiation
- PMID: 35163045
- PMCID: PMC8834919
- DOI: 10.3390/ijms23031115
Temporal Gene Expression Profiles Reflect the Dynamics of Lymphoid Differentiation
Abstract
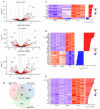
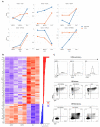
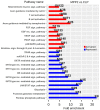
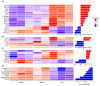
Understanding the emergence of lymphoid committed cells from multipotent progenitors (MPP) is a great challenge in hematopoiesis. To gain deeper insight into the dynamic expression changes associated with these transitions, we report the quantitative transcriptome of two MPP subsets and the common lymphoid progenitor (CLP). While the transcriptome is rather stable between MPP2 and MPP3, expression changes increase with differentiation. Among those, we found that pioneer lymphoid genes such as Rag1, Mpeg1, and Dntt are expressed continuously from MPP2. Others, such as CD93, are CLP specific, suggesting their potential use as new markers to improve purification of lymphoid populations. Notably, a six-transcription factor network orchestrates the lymphoid differentiation program. Additionally, we pinpointed 24 long intergenic-non-coding RNA (lincRNA) differentially expressed through commitment and further identified seven novel forms. Collectively, our approach provides a comprehensive landscape of coding and non-coding transcriptomes expressed during lymphoid commitment.
Keywords: RNAseq; gene networks; hematopoietic differentiation; long non-coding RNA; lymphopoiesis; transcriptome.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


Similar articles
-
Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages.Nat Immunol. 2015 Dec;16(12):1282-91. doi: 10.1038/ni.3299. Epub 2015 Oct 26. Nat Immunol. 2015. PMID: 26502406 Free PMC article.
-
Long noncoding RNAs of single hematopoietic stem and progenitor cells in healthy and dysplastic human bone marrow.Haematologica. 2019 May;104(5):894-906. doi: 10.3324/haematol.2018.208926. Epub 2018 Dec 13. Haematologica. 2019. PMID: 30545929 Free PMC article.
-
Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing.Mol Vis. 2014 Nov 4;20:1491-517. eCollection 2014. Mol Vis. 2014. PMID: 25489224 Free PMC article.
-
Pbx1 restrains myeloid maturation while preserving lymphoid potential in hematopoietic progenitors.J Cell Sci. 2013 Jul 15;126(Pt 14):3181-91. doi: 10.1242/jcs.125435. Epub 2013 May 9. J Cell Sci. 2013. PMID: 23660001 Free PMC article.
-
Critical differences in hematopoiesis and lymphoid development between humans and mice.J Clin Immunol. 2013 May;33(4):711-5. doi: 10.1007/s10875-012-9844-3. Epub 2012 Dec 30. J Clin Immunol. 2013. PMID: 23274800 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

