Three-axis classification of mouse lung mesenchymal cells reveals two populations of myofibroblasts
- PMID: 35302583
- PMCID: PMC8977099
- DOI: 10.1242/dev.200081
Three-axis classification of mouse lung mesenchymal cells reveals two populations of myofibroblasts
Abstract
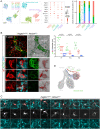
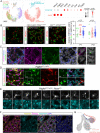
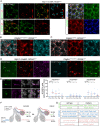
The mesenchyme consists of heterogeneous cell populations that support neighboring structures and are integral to intercellular signaling, but are poorly defined morphologically and molecularly. Leveraging single-cell RNA-sequencing, 3D imaging and lineage tracing, we classify the mouse lung mesenchyme into three proximal-distal axes that are associated with the endothelium, epithelium and interstitium, respectively. From proximal to distal: the vascular axis includes vascular smooth muscle cells and pericytes that transition as arterioles and venules ramify into capillaries; the epithelial axis includes airway smooth muscle cells and two populations of myofibroblasts - ductal myofibroblasts, surrounding alveolar ducts and marked by CDH4, HHIP and LGR6, which persist post-alveologenesis, and alveolar myofibroblasts, surrounding alveoli and marked by high expression of PDGFRA, which undergo developmental apoptosis; and the interstitial axis, residing between the epithelial and vascular trees and sharing the marker MEOX2, includes fibroblasts in the bronchovascular bundle and the alveolar interstitium, which are marked by IL33/DNER/PI16 and Wnt2, respectively. Single-cell imaging reveals a distinct morphology of mesenchymal cell populations. This classification provides a conceptual and experimental framework applicable to other organs.
Keywords: 3D imaging; Lung development; Mesenchymal cells; Mouse; Single-cell genomics.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




Similar articles
-
A three-dimensional study of alveologenesis in mouse lung.Dev Biol. 2016 Jan 15;409(2):429-41. doi: 10.1016/j.ydbio.2015.11.017. Epub 2015 Nov 26. Dev Biol. 2016. PMID: 26632490 Free PMC article.
-
Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6.Cell. 2017 Sep 7;170(6):1149-1163.e12. doi: 10.1016/j.cell.2017.07.028. Cell. 2017. PMID: 28886383 Free PMC article.
-
Fgf10-positive cells represent a progenitor cell population during lung development and postnatally.Development. 2014 Jan;141(2):296-306. doi: 10.1242/dev.099747. Epub 2013 Dec 18. Development. 2014. PMID: 24353064 Free PMC article.
-
Alveologenesis: What Governs Secondary Septa Formation.Int J Mol Sci. 2021 Nov 9;22(22):12107. doi: 10.3390/ijms222212107. Int J Mol Sci. 2021. PMID: 34829987 Free PMC article. Review.
-
Paracrine cellular and extracellular matrix interactions with mesenchymal progenitors during pulmonary alveolar septation.Birth Defects Res A Clin Mol Teratol. 2014 Mar;100(3):227-39. doi: 10.1002/bdra.23230. Epub 2014 Mar 17. Birth Defects Res A Clin Mol Teratol. 2014. PMID: 24639378 Review.
Cited by
-
Hyperoxia prevents the dynamic neonatal increases in lung mesenchymal cell diversity.Sci Rep. 2024 Jan 23;14(1):2033. doi: 10.1038/s41598-023-50717-w. Sci Rep. 2024. PMID: 38263350 Free PMC article.
-
Impaired Myofibroblast Proliferation is a Central Feature of Pathologic Post-Natal Alveolar Simplification.bioRxiv [Preprint]. 2024 Sep 16:2023.12.21.572766. doi: 10.1101/2023.12.21.572766. bioRxiv. 2024. Update in: Elife. 2024 Dec 11;13:RP94425. doi: 10.7554/eLife.94425 PMID: 38187712 Free PMC article. Updated. Preprint.
-
Hedgehog and Platelet-derived Growth Factor Signaling Intersect during Postnatal Lung Development.Am J Respir Cell Mol Biol. 2023 May;68(5):523-536. doi: 10.1165/rcmb.2022-0269OC. Am J Respir Cell Mol Biol. 2023. PMID: 36693140 Free PMC article.
-
Alveolar fibroblast lineage orchestrates lung inflammation and fibrosis.Nature. 2024 Jul;631(8021):627-634. doi: 10.1038/s41586-024-07660-1. Epub 2024 Jul 10. Nature. 2024. PMID: 38987592
-
Mitochondrial integrated stress response controls lung epithelial cell fate.Nature. 2023 Aug;620(7975):890-897. doi: 10.1038/s41586-023-06423-8. Epub 2023 Aug 9. Nature. 2023. PMID: 37558881 Free PMC article.
References
-
- Cassandras, M., Wang, C., Kathiriya, J., Tsukui, T., Matatia, P., Matthay, M., Wolters, P., Molofsky, A., Sheppard, D., Chapman, H.et al. (2020). Gli1(+) mesenchymal stromal cells form a pathological niche to promote airway progenitor metaplasia in the fibrotic lung. Nat. Cell Biol. 22, 1295-1306. 10.1038/s41556-020-00591-9 - DOI - PMC - PubMed
-
- Dahlgren, M. W., Jones, S. W., Cautivo, K. M., Dubinin, A., Ortiz-Carpena, J. F., Farhat, S., Yu, K. S., Lee, K., Wang, C., Molofsky, A. V.et al. (2019). Adventitial stromal cells define Group 2 innate lymphoid cell tissue niches. Immunity 50, 707-722. 10.1016/j.immuni.2019.02.002 - DOI - PMC - PubMed
-
- El Agha, E., Herold, S., Al Alam, D., Quantius, J., MacKenzie, B., Carraro, G., Moiseenko, A., Chao, C. M., Minoo, P., Seeger, W.et al. (2014). Fgf10-positive cells represent a progenitor cell population during lung development and postnatally. Development 141, 296-306. 10.1242/dev.099747 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

