Lysine methyltransferase inhibitors: where we are now
- PMID: 35441141
- PMCID: PMC8985178
- DOI: 10.1039/d1cb00196e
Lysine methyltransferase inhibitors: where we are now
Abstract
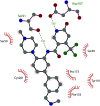
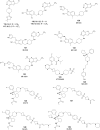
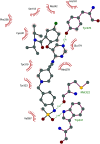
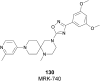
Protein lysine methyltransferases constitute a large family of epigenetic writers that catalyse the transfer of a methyl group from the cofactor S-adenosyl-l-methionine to histone- and non-histone-specific substrates. Alterations in the expression and activity of these proteins have been linked to the genesis and progress of several diseases, including cancer, neurological disorders, and growing defects, hence they represent interesting _targets for new therapeutic approaches. Over the past two decades, the identification of modulators of lysine methyltransferases has increased tremendously, clarifying the role of these proteins in different physio-pathological states. The aim of this review is to furnish an updated outlook about the protein lysine methyltransferases disclosed modulators, reporting their potency, their mechanism of action and their eventual use in clinical and preclinical studies.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
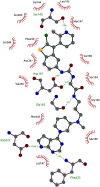
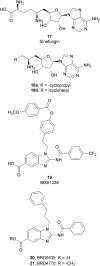
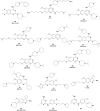
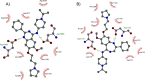
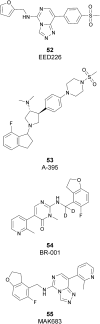
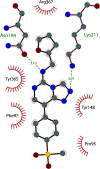
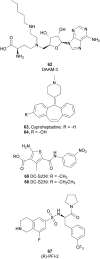






Similar articles
-
Protein lysine methylation by seven-β-strand methyltransferases.Biochem J. 2016 Jul 15;473(14):1995-2009. doi: 10.1042/BCJ20160117. Biochem J. 2016. PMID: 27407169 Review.
-
A simplified characterization of S-adenosyl-l-methionine-consuming enzymes with 1-Step EZ-MTase: a universal and straightforward coupled-assay for in vitro and in vivo setting.Chem Sci. 2017 Sep 1;8(9):6601-6612. doi: 10.1039/c7sc02830j. Epub 2017 Jul 27. Chem Sci. 2017. PMID: 29449933 Free PMC article.
-
Synthesis of lysine methyltransferase inhibitors.Front Chem. 2015 Jul 23;3:44. doi: 10.3389/fchem.2015.00044. eCollection 2015. Front Chem. 2015. PMID: 26258118 Free PMC article. Review.
-
Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases.J Biol Chem. 2006 Jul 14;281(28):19280-7. doi: 10.1074/jbc.M602257200. Epub 2006 May 8. J Biol Chem. 2006. PMID: 16682405
-
S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.Proc Natl Acad Sci U S A. 2016 May 31;113(22):6182-7. doi: 10.1073/pnas.1605523113. Epub 2016 May 16. Proc Natl Acad Sci U S A. 2016. PMID: 27185940 Free PMC article.
Cited by
-
Lead-oriented synthesis of epigenetic relevant scaffolds.Chem Commun (Camb). 2023 Dec 7;59(98):14555-14558. doi: 10.1039/d3cc04317g. Chem Commun (Camb). 2023. PMID: 37991354 Free PMC article.
-
Turning Nonselective Inhibitors of Type I Protein Arginine Methyltransferases into Potent and Selective Inhibitors of Protein Arginine Methyltransferase 4 through a Deconstruction-Reconstruction and Fragment-Growing Approach.J Med Chem. 2022 Sep 8;65(17):11574-11606. doi: 10.1021/acs.jmedchem.2c00252. Epub 2022 Apr 28. J Med Chem. 2022. PMID: 35482954 Free PMC article.
-
Euchromatic Histone Lysine Methyltransferase 2 Inhibition Enhances Carfilzomib Sensitivity and Overcomes Drug Resistance in Multiple Myeloma Cell Lines.Cancers (Basel). 2023 Apr 7;15(8):2199. doi: 10.3390/cancers15082199. Cancers (Basel). 2023. PMID: 37190128 Free PMC article.
-
Enzymatic synthesis of S-adenosyl-l-homocysteine and its nucleoside analogs from racemic homocysteine thiolactone.Chem Sci. 2024 Sep 6;15(38):15900-6. doi: 10.1039/d4sc03801k. Online ahead of print. Chem Sci. 2024. PMID: 39282651 Free PMC article.
-
Impact of Histone Lysine Methyltransferase SUV4-20H2 on Cancer Onset and Progression with Therapeutic Potential.Int J Mol Sci. 2024 Feb 21;25(5):2498. doi: 10.3390/ijms25052498. Int J Mol Sci. 2024. PMID: 38473745 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

