Organismal and Cellular Stress Responses upon Disruption of Mitochondrial Lonp1 Protease
- PMID: 35456042
- PMCID: PMC9025075
- DOI: 10.3390/cells11081363
Organismal and Cellular Stress Responses upon Disruption of Mitochondrial Lonp1 Protease
Abstract
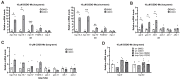
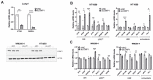
Cells engage complex surveillance mechanisms to maintain mitochondrial function and protein homeostasis. LonP1 protease is a key component of mitochondrial quality control and has been implicated in human malignancies and other pathological disorders. Here, we employed two experimental systems, the worm Caenorhabditis elegans and human cancer cells, to investigate and compare the effects of LONP-1/LonP1 deficiency at the molecular, cellular, and organismal levels. Deletion of the lonp-1 gene in worms disturbed mitochondrial function, provoked reactive oxygen species accumulation, and impaired normal processes, such as growth, behavior, and lifespan. The viability of lonp-1 mutants was dependent on the activity of the ATFS-1 transcription factor, and loss of LONP-1 evoked retrograde signaling that involved both the mitochondrial and cytoplasmic unfolded protein response (UPRmt and UPRcyt) pathways and ensuing diverse organismal stress responses. Exposure of worms to triterpenoid CDDO-Me, an inhibitor of human LonP1, stimulated only UPRcyt responses. In cancer cells, CDDO-Me induced key components of the integrated stress response (ISR), the UPRmt and UPRcyt pathways, and the redox machinery. However, genetic knockdown of LonP1 revealed a genotype-specific cellular response and induced apoptosis similar to CDDO-Me treatment. Overall, the mitochondrial dysfunction ensued by disruption of LonP1 elicits adaptive cytoprotective mechanisms that can inhibit cancer cell survival but diversely modulate organismal stress response and aging.
Keywords: C. elegans; CDDO-Me; LonP1; aging; cancer; mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



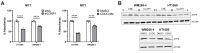
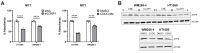
Similar articles
-
Mitochondrial p38 Mitogen-Activated Protein Kinase: Insights into Its Regulation of and Role in LONP1-Deficient Nematodes.Int J Mol Sci. 2023 Dec 7;24(24):17209. doi: 10.3390/ijms242417209. Int J Mol Sci. 2023. PMID: 38139038 Free PMC article.
-
Inhibition of mitochondrial LonP1 protease by allosteric blockade of ATP binding and hydrolysis via CDDO and its derivatives.J Biol Chem. 2022 Mar;298(3):101719. doi: 10.1016/j.jbc.2022.101719. Epub 2022 Feb 11. J Biol Chem. 2022. PMID: 35151690 Free PMC article.
-
CDDO-Me Selectively Attenuates CA1 Neuronal Death Induced by Status Epilepticus via Facilitating Mitochondrial Fission Independent of LONP1.Cells. 2019 Aug 5;8(8):833. doi: 10.3390/cells8080833. Cells. 2019. PMID: 31387295 Free PMC article.
-
The mitochondrial unfolded protein response: A multitasking giant in the fight against human diseases.Ageing Res Rev. 2022 Nov;81:101702. doi: 10.1016/j.arr.2022.101702. Epub 2022 Jul 28. Ageing Res Rev. 2022. PMID: 35908669 Review.
-
Mitochondrial recovery by the UPRmt: Insights from C. elegans.Semin Cell Dev Biol. 2024 Feb 15;154(Pt A):59-68. doi: 10.1016/j.semcdb.2023.02.002. Epub 2023 Feb 13. Semin Cell Dev Biol. 2024. PMID: 36792440 Free PMC article. Review.
Cited by
-
Mitochondrial p38 Mitogen-Activated Protein Kinase: Insights into Its Regulation of and Role in LONP1-Deficient Nematodes.Int J Mol Sci. 2023 Dec 7;24(24):17209. doi: 10.3390/ijms242417209. Int J Mol Sci. 2023. PMID: 38139038 Free PMC article.
-
A bird's eye view of mitochondrial unfolded protein response in cancer: mechanisms, progression and further applications.Cell Death Dis. 2024 Sep 11;15(9):667. doi: 10.1038/s41419-024-07049-y. Cell Death Dis. 2024. PMID: 39261452 Free PMC article. Review.
-
Hypoxia-induced mitochondrial stress granules.Cell Death Dis. 2023 Jul 19;14(7):448. doi: 10.1038/s41419-023-05988-6. Cell Death Dis. 2023. PMID: 37468471 Free PMC article.
-
The synthetic oleanane triterpenoid CDDO-2P-Im binds GRP78/BiP to induce unfolded protein response-mediated apoptosis in myeloma.Mol Oncol. 2023 Dec;17(12):2526-2545. doi: 10.1002/1878-0261.13447. Epub 2023 Jun 13. Mol Oncol. 2023. PMID: 37149844 Free PMC article.
-
The Role of Lonp1 on Mitochondrial Functions during Cardiovascular and Muscular Diseases.Antioxidants (Basel). 2023 Feb 28;12(3):598. doi: 10.3390/antiox12030598. Antioxidants (Basel). 2023. PMID: 36978846 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

