Comparative transcriptome and metabolome analyses reveal the methanol dissimilation pathway of Pichia pastoris
- PMID: 35549850
- PMCID: PMC9103059
- DOI: 10.1186/s12864-022-08592-8
Comparative transcriptome and metabolome analyses reveal the methanol dissimilation pathway of Pichia pastoris
Abstract
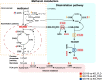
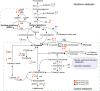
Background: Pichia pastoris (Komagataella phaffii) is a model organism widely used for the recombinant expression of eukaryotic proteins, and it can metabolize methanol as its sole carbon and energy source. Methanol is oxidized to formaldehyde by alcohol oxidase (AOX). In the dissimilation pathway, formaldehyde is oxidized to CO2 by formaldehyde dehydrogenase (FLD), S-hydroxymethyl glutathione hydrolase (FGH) and formate dehydrogenase (FDH).
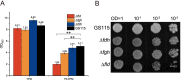
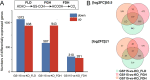
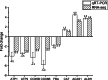
Results: The transcriptome and metabolome of P. pastoris were determined under methanol cultivation when its dissimilation pathway cut off. Firstly, Δfld and Δfgh were significantly different compared to the wild type (GS115), with a 60.98% and 23.66% reduction in biomass, respectively. The differential metabolites between GS115 and Δfld were mainly enriched in ABC transporters, amino acid biosynthesis, and protein digestion and absorption. Secondly, comparative transcriptome between knockout and wild type strains showed that oxidative phosphorylation, glycolysis and the TCA cycle were downregulated, while alcohol metabolism, proteasomes, autophagy and peroxisomes were upregulated. Interestingly, the down-regulation of the oxidative phosphorylation pathway was positively correlated with the gene order of dissimilation pathway knockdown. In addition, there were significant differences in amino acid metabolism and glutathione redox cycling that raised our concerns about formaldehyde sorption in cells.
Conclusions: This is the first time that integrity of dissimilation pathway analysis based on transcriptomics and metabolomics was carried out in Pichia pastoris. The blockage of dissimilation pathway significantly down-regulates the level of oxidative phosphorylation and weakens the methanol assimilation pathway to the point where deficiencies in energy supply and carbon fixation result in inefficient biomass accumulation and genetic replication. In addition, transcriptional upregulation of the proteasome and autophagy may be a stress response to resolve formaldehyde-induced DNA-protein crosslinking.
Keywords: Dissimilation pathway; Formaldehyde; Glutathione; Methanol metabolism; Oxidative phosphorylation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Metabolic engineering of Komagataella phaffii for the efficient utilization of methanol.Microb Cell Fact. 2024 Jul 17;23(1):198. doi: 10.1186/s12934-024-02475-1. Microb Cell Fact. 2024. PMID: 39014373 Free PMC article.
-
The Mut+ strain of Komagataella phaffii (Pichia pastoris) expresses PAOX1 5 and 10 times faster than Muts and Mut- strains: evidence that formaldehyde or/and formate are true inducers of PAOX1.Appl Microbiol Biotechnol. 2020 Sep;104(18):7801-7814. doi: 10.1007/s00253-020-10793-8. Epub 2020 Aug 6. Appl Microbiol Biotechnol. 2020. PMID: 32761464
-
Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes.Microb Cell Fact. 2012 Aug 8;11:103. doi: 10.1186/1475-2859-11-103. Microb Cell Fact. 2012. PMID: 22873405 Free PMC article.
-
Microbe Profile: Komagataella phaffii: a methanol devouring biotech yeast formerly known as Pichia pastoris.Microbiology (Reading). 2020 Jul;166(7):614-616. doi: 10.1099/mic.0.000958. Microbiology (Reading). 2020. PMID: 32720891 Review.
-
The significance of peroxisomes in methanol metabolism in methylotrophic yeast.Biochim Biophys Acta. 2006 Dec;1763(12):1453-62. doi: 10.1016/j.bbamcr.2006.07.016. Epub 2006 Sep 1. Biochim Biophys Acta. 2006. PMID: 17023065 Review.
Cited by
-
Hansenula polymorpha methanol metabolism genes enhance recombinant protein production in Komagataella phaffi.AMB Express. 2024 Aug 2;14(1):88. doi: 10.1186/s13568-024-01743-y. AMB Express. 2024. PMID: 39095661 Free PMC article.
-
Metabolic engineering of Komagataella phaffii for the efficient utilization of methanol.Microb Cell Fact. 2024 Jul 17;23(1):198. doi: 10.1186/s12934-024-02475-1. Microb Cell Fact. 2024. PMID: 39014373 Free PMC article.
-
What makes Candida auris pan-drug resistant? Integrative insights from genomic, transcriptomic, and phenomic analysis of clinical strains resistant to all four major classes of antifungal drugs.Antimicrob Agents Chemother. 2024 Oct 8;68(10):e0091124. doi: 10.1128/aac.00911-24. Epub 2024 Sep 19. Antimicrob Agents Chemother. 2024. PMID: 39297640
-
Engineering Komagataella phaffii to biosynthesize cordycepin from methanol which drives global metabolic alterations at the transcription level.Synth Syst Biotechnol. 2023 Mar 16;8(2):242-252. doi: 10.1016/j.synbio.2023.03.003. eCollection 2023 Jun. Synth Syst Biotechnol. 2023. PMID: 37007278 Free PMC article.
-
Alternative PCR-Based Approaches for Generation of Komagataella phaffii Strains.Microorganisms. 2023 Sep 12;11(9):2297. doi: 10.3390/microorganisms11092297. Microorganisms. 2023. PMID: 37764140 Free PMC article.
References
-
- Johnson MA, Waterham HR, Ksheminska GP, Fayura LR, Cereghino JL, Stasyk OV, Veenhuis M, Kulachkovsky AR, Sibirny AA, Cregg JM. Positive Selection of Novel Peroxisome Biogenesis-Defective Mutants of the Yeast Pichia pastoris. Genetics. 1999;151(4):1379–1391. doi: 10.1093/genetics/151.4.1379. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

