USP16-mediated histone H2A lysine-119 deubiquitination during oocyte maturation is a prerequisite for zygotic genome activation
- PMID: 35640597
- PMCID: PMC9178006
- DOI: 10.1093/nar/gkac468
USP16-mediated histone H2A lysine-119 deubiquitination during oocyte maturation is a prerequisite for zygotic genome activation
Abstract
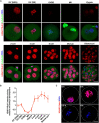
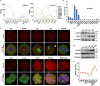
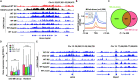
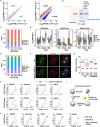
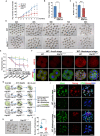
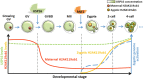
Maternal-to-zygotic transition (MZT) is the first and key step in the control of animal development and intimately related to changes in chromatin structure and histone modifications. H2AK119ub1, an important epigenetic modification in regulating chromatin configuration and function, is primarily catalyzed by PRC1 and contributes to resistance to transcriptional reprogramming in mouse embryos. In this study, the genome-wide dynamic distribution of H2AK119ub1 during MZT in mice was investigated using chromosome immunoprecipitation and sequencing. The results indicated that H2AK119ub1 accumulated in fully grown oocytes and was enriched at the TSSs of maternal genes, but was promptly declined after meiotic resumption at genome-wide including the TSSs of early zygotic genes, by a previously unidentified mechanism. Genetic evidences indicated that ubiquitin-specific peptidase 16 (USP16) is the major deubiquitinase for H2AK119ub1 in mouse oocytes. Conditional knockout of Usp16 in oocytes did not impair their survival, growth, or meiotic maturation. However, oocytes lacking USP16 have defects when undergoing zygotic genome activation or gaining developmental competence after fertilization, potentially associated with high levels of maternal H2AK119ub1 deposition on the zygotic genomes. Taken together, H2AK119ub1 level is declined during oocyte maturation by an USP16-dependent mechanism, which ensures zygotic genome reprogramming and transcriptional activation of essential early zygotic genes.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition.Nature. 2016 Sep 22;537(7621):548-552. doi: 10.1038/nature19360. Epub 2016 Sep 14. Nature. 2016. PMID: 27626377 Free PMC article.
-
H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos.Nat Genet. 2021 Apr;53(4):539-550. doi: 10.1038/s41588-021-00820-3. Epub 2021 Apr 5. Nat Genet. 2021. PMID: 33821003
-
Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos.Nat Genet. 2021 Apr;53(4):551-563. doi: 10.1038/s41588-021-00821-2. Epub 2021 Apr 5. Nat Genet. 2021. PMID: 33821005 Free PMC article.
-
Function and Regulation of Histone H3 Lysine-4 Methylation During Oocyte Meiosis and Maternal-to-Zygotic Transition.Front Cell Dev Biol. 2020 Oct 9;8:597498. doi: 10.3389/fcell.2020.597498. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33163498 Free PMC article. Review.
-
A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammals†.Biol Reprod. 2019 Sep 1;101(3):579-590. doi: 10.1093/biolre/ioz012. Biol Reprod. 2019. PMID: 30715134 Review.
Cited by
-
Epigenetic reprogramming during the maternal-to-zygotic transition.MedComm (2020). 2023 Aug 2;4(4):e331. doi: 10.1002/mco2.331. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37547174 Free PMC article. Review.
-
Know when to fold 'em: Polycomb complexes in oncogenic 3D genome regulation.Front Cell Dev Biol. 2022 Aug 29;10:986319. doi: 10.3389/fcell.2022.986319. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36105358 Free PMC article. Review.
-
DNA Damage Induction Alters the Expression of Ubiquitin and SUMO Regulators in Preimplantation Stage Pig Embryos.Int J Mol Sci. 2022 Aug 25;23(17):9610. doi: 10.3390/ijms23179610. Int J Mol Sci. 2022. PMID: 36077022 Free PMC article.
-
The Pleiotropic Ubiquitin-Specific Peptidase 16 and Its Many Substrates.Cells. 2023 Mar 13;12(6):886. doi: 10.3390/cells12060886. Cells. 2023. PMID: 36980227 Free PMC article. Review.
-
Current status of genome-wide epigenetic profiling of mammalian preimplantation embryos.Reprod Med Biol. 2023 Jun 21;22(1):e12521. doi: 10.1002/rmb2.12521. eCollection 2023 Jan-Dec. Reprod Med Biol. 2023. PMID: 37351110 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

