Sleep Loss Causes Dysfunction in Murine Extraorbital Lacrimal Glands
- PMID: 35731510
- PMCID: PMC9233287
- DOI: 10.1167/iovs.63.6.19
Sleep Loss Causes Dysfunction in Murine Extraorbital Lacrimal Glands
Erratum in
-
Erratum in: Sleep Loss Causes Dysfunction in Murine Extraorbital Lacrimal Glands.Invest Ophthalmol Vis Sci. 2022 Jul 8;63(8):13. doi: 10.1167/iovs.63.8.13. Invest Ophthalmol Vis Sci. 2022. PMID: 35822951 Free PMC article. No abstract available.
Abstract
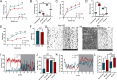
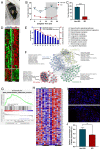
Purpose: Sleep loss markedly affects the structure and function of the lacrimal gland and may cause ocular surface disease as a common public health problem. This study aims to investigate the circadian disturbance caused by sleep loss leading to dysfunction of extraorbital lacrimal glands (ELGs).
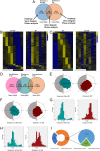
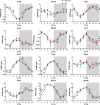
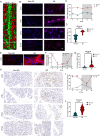
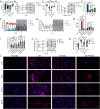
Methods: A mouse sleep deprivation (SD) model for sleep loss studies was built in C57BL/6J male mice. After four weeks, the ELGs were collected at three-hour intervals during a 24-hour period. The Jonckheere-Terpstra-Kendall algorithm was used to determine the composition, phase, and rhythmicity of transcriptomic profiles in ELGs. Furthermore, we compared the non-sleep-deprived and SD-treated mouse ELG (i) reactive oxygen species (ROS) by fluorescein staining, (ii) DNA damage by immunostaining for γ-H2Ax, and (iii) circadian migration of immune cells by immunostaining for CD4, CD8, γδ-TCR, CD64, and CX3CR1. Finally, we also evaluated (i) the locomotor activity and core body temperature rhythm of mice and (ii) the mass, cell size, and tear secretion of the ELGs.
Results: SD dramatically altered the composition and phase-associated functional enrichment of the circadian transcriptome, immune cell trafficking, metabolism, cell differentiation, and neural secretory activities of mouse ELGs. Additionally, SD caused the ROS accumulation and consequent DNA damage in the ELGs, and the ELG dysfunction caused by SD was irreversible.
Conclusions: SD damages the structure, function, and diurnal oscillations of ELGs. These results highlight comprehensive characterization of insufficient sleep-affected ELG circadian transcriptome that may provide a new therapeutic approach to counteract the effects of SD on ELG function.
Conflict of interest statement
Disclosure:
Figures



Similar articles
-
Type 1 diabetes mellitus impairs diurnal oscillations in murine extraorbital lacrimal glands.Ocul Surf. 2020 Jul;18(3):438-452. doi: 10.1016/j.jtos.2020.04.013. Epub 2020 May 1. Ocul Surf. 2020. PMID: 32360784
-
High-Fat Nutritional Challenge Reshapes Circadian Signatures in Murine Extraorbital Lacrimal Glands.Invest Ophthalmol Vis Sci. 2022 May 2;63(5):23. doi: 10.1167/iovs.63.5.23. Invest Ophthalmol Vis Sci. 2022. PMID: 35588356 Free PMC article.
-
Light cycle phase advance as a model for jet lag reprograms the circadian rhythms of murine extraorbital lacrimal glands.Ocul Surf. 2021 Apr;20:95-114. doi: 10.1016/j.jtos.2021.02.001. Epub 2021 Feb 11. Ocul Surf. 2021. PMID: 33582293
-
A new mouse model of dry eye disease: oxidative stress affects functional decline in the lacrimal gland.Cornea. 2012 Nov;31 Suppl 1:S63-7. doi: 10.1097/ICO.0b013e31826a5de1. Cornea. 2012. PMID: 23038038 Review.
-
How sleep and wakefulness influence circadian rhythmicity: effects of insufficient and mistimed sleep on the animal and human transcriptome.J Sleep Res. 2015 Oct;24(5):476-93. doi: 10.1111/jsr.12307. Epub 2015 Jun 8. J Sleep Res. 2015. PMID: 26059855 Review.
Cited by
-
High-fat intake reshapes the circadian transcriptome profile and metabolism in murine meibomian glands.Front Nutr. 2023 Mar 16;10:1146916. doi: 10.3389/fnut.2023.1146916. eCollection 2023. Front Nutr. 2023. PMID: 37006922 Free PMC article.
-
Mechanisms of Extraorbital Lacrimal Gland Aging in Mice: An Integrative Analysis of the Temporal Transcriptome.Invest Ophthalmol Vis Sci. 2023 Sep 1;64(12):18. doi: 10.1167/iovs.64.12.18. Invest Ophthalmol Vis Sci. 2023. PMID: 37695604 Free PMC article.
-
Enterovirus A71 infection-induced dry eye-like symptoms by damaging the lacrimal glands.Front Cell Infect Microbiol. 2024 Apr 2;14:1340075. doi: 10.3389/fcimb.2024.1340075. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38628549 Free PMC article.
-
Diabetes Reshapes the Circadian Transcriptome Profile in Murine Retina.Invest Ophthalmol Vis Sci. 2023 Oct 3;64(13):3. doi: 10.1167/iovs.64.13.3. Invest Ophthalmol Vis Sci. 2023. PMID: 37788001 Free PMC article.
-
Sleep deprivation induces corneal endothelial dysfunction by downregulating Bmal1.BMC Ophthalmol. 2024 Jun 21;24(1):268. doi: 10.1186/s12886-024-03524-4. BMC Ophthalmol. 2024. PMID: 38907352 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

