Lipolysis regulates major transcriptional programs in brown adipocytes
- PMID: 35803907
- PMCID: PMC9270495
- DOI: 10.1038/s41467-022-31525-8
Lipolysis regulates major transcriptional programs in brown adipocytes
Abstract
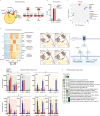
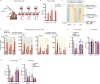
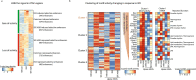
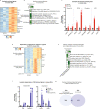
β-Adrenergic signaling is a core regulator of brown adipocyte function stimulating both lipolysis and transcription of thermogenic genes, thereby expanding the capacity for oxidative metabolism. We have used pharmacological inhibitors and a direct activator of lipolysis to acutely modulate the activity of lipases, thereby enabling us to uncover lipolysis-dependent signaling pathways downstream of β-adrenergic signaling in cultured brown adipocytes. Here we show that induction of lipolysis leads to acute induction of several gene programs and is required for transcriptional regulation by β-adrenergic signals. Using machine-learning algorithms to infer causal transcription factors, we show that PPARs are key mediators of lipolysis-induced activation of genes involved in lipid metabolism and thermogenesis. Importantly, however, lipolysis also activates the unfolded protein response and regulates the core circadian transcriptional machinery independently of PPARs. Our results demonstrate that lipolysis generates important metabolic signals that exert profound pleiotropic effects on transcription and function of cultured brown adipocytes.
© 2022. The Author(s).
Conflict of interest statement
O.S.J and Z.G.H work or have worked, in some capacity, for Embark Biotech ApS, a company developing therapeutics for the treatment of diabetes and obesity. The remaining authors declare no competing interests.
Figures



Similar articles
-
Adipose retinol saturase is regulated by β-adrenergic signaling and its deletion impairs lipolysis in adipocytes and acute cold tolerance in mice.Mol Metab. 2024 Jan;79:101855. doi: 10.1016/j.molmet.2023.101855. Epub 2023 Dec 19. Mol Metab. 2024. PMID: 38128827 Free PMC article.
-
β3-Adrenergic receptors regulate human brown/beige adipocyte lipolysis and thermogenesis.JCI Insight. 2021 Jun 8;6(11):e139160. doi: 10.1172/jci.insight.139160. JCI Insight. 2021. PMID: 34100382 Free PMC article.
-
Adaptive thermogenesis in brown adipose tissue involves activation of pannexin-1 channels.Mol Metab. 2021 Feb;44:101130. doi: 10.1016/j.molmet.2020.101130. Epub 2020 Nov 25. Mol Metab. 2021. PMID: 33248294 Free PMC article.
-
Positive and negative control of Ucp1 gene transcription and the role of β-adrenergic signaling networks.Int J Obes (Lond). 2010 Oct;34 Suppl 1:S28-33. doi: 10.1038/ijo.2010.180. Int J Obes (Lond). 2010. PMID: 20935662 Review.
-
Second messenger signaling mechanisms of the brown adipocyte thermogenic program: an integrative perspective.Horm Mol Biol Clin Investig. 2017 Sep 26;31(2):/j/hmbci.2017.31.issue-2/hmbci-2017-0062/hmbci-2017-0062.xml. doi: 10.1515/hmbci-2017-0062. Horm Mol Biol Clin Investig. 2017. PMID: 28949928 Review.
Cited by
-
p21-activated kinase 4 counteracts PKA-dependent lipolysis by phosphorylating FABP4 and HSL.Nat Metab. 2024 Jan;6(1):94-112. doi: 10.1038/s42255-023-00957-x. Epub 2024 Jan 12. Nat Metab. 2024. PMID: 38216738
-
Extract mixture of plants (OXYLIA) inhibits fat accumulation by blocking FAS-related factors and promoting lipolysis via cAMP-dependent PKA activation.Food Nutr Res. 2024 Mar 12;68. doi: 10.29219/fnr.v68.10180. eCollection 2024. Food Nutr Res. 2024. PMID: 38571921 Free PMC article.
-
Adipose tissue at single-cell resolution.Cell Metab. 2023 Mar 7;35(3):386-413. doi: 10.1016/j.cmet.2023.02.002. Cell Metab. 2023. PMID: 36889280 Free PMC article. Review.
-
Creatine and low-dose lithium supplementation separately alter energy expenditure, body mass, and adipose metabolism for the promotion of thermogenesis.iScience. 2024 Mar 11;27(4):109468. doi: 10.1016/j.isci.2024.109468. eCollection 2024 Apr 19. iScience. 2024. PMID: 38550985 Free PMC article.
-
RNA-Seq Study on the Longissimus thoracis Muscle of Italian Large White Pigs Fed Extruded Linseed with or without Antioxidants and Polyphenols.Animals (Basel). 2023 Mar 28;13(7):1187. doi: 10.3390/ani13071187. Animals (Basel). 2023. PMID: 37048443 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

