The Oral Microbiome and Its Role in Systemic Autoimmune Diseases: A Systematic Review of Big Data Analysis
- PMID: 35844967
- PMCID: PMC9277227
- DOI: 10.3389/fdata.2022.927520
The Oral Microbiome and Its Role in Systemic Autoimmune Diseases: A Systematic Review of Big Data Analysis
Abstract
Introduction: Despite decades of research, systemic autoimmune diseases (SADs) continue to be a major global health concern and the etiology of these diseases is still not clear. To date, with the development of high-throughput techniques, increasing evidence indicated a key role of oral microbiome in the pathogenesis of SADs, and the alterations of oral microbiome may contribute to the disease emergence or evolution. This review is to present the latest knowledge on the relationship between the oral microbiome and SADs, focusing on the multiomics data generated from a large set of samples.
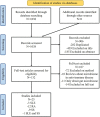
Methodology: By searching the PubMed and Embase databases, studies that investigated the oral microbiome of SADs, including systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and Sjögren's syndrome (SS), were systematically reviewed according to the PRISMA guidelines.
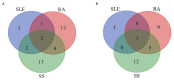
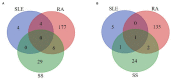
Results: One thousand and thirty-eight studies were found, and 25 studies were included: three referred to SLE, 12 referred to RA, nine referred to SS, and one to both SLE and SS. The 16S rRNA sequencing was the most frequent technique used. HOMD was the most common database aligned to and QIIME was the most popular pipeline for downstream analysis. Alterations in bacterial composition and population have been found in the oral samples of patients with SAD compared with the healthy controls. Results regarding candidate pathogens were not always in accordance, but Selenomonas and Veillonella were found significantly increased in three SADs, and Streptococcus was significantly decreased in the SADs compared with controls.
Conclusion: A large amount of sequencing data was collected from patients with SAD and controls in this systematic review. Oral microbial dysbiosis had been identified in these SADs, although the dysbiosis features were different among studies. There was a lack of standardized study methodology for each study from the inclusion criteria, sample type, sequencing platform, and referred database to downstream analysis pipeline and cutoff. Besides the genomics, transcriptomics, proteomics, and metabolomics technology should be used to investigate the oral microbiome of patients with SADs and also the at-risk individuals of disease development, which may provide us with a better understanding of the etiology of SADs and promote the development of the novel therapies.
Keywords: Sjögren's syndrome; high-throughput analysis; oral microbiome; rheumatoid arthritis; systemic autoimmune disease; systemic lupus erythematosus.
Copyright © 2022 Gao, Cheng, Zhu, Bi, Shi and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The autoimmunity-oral microbiome connection.Oral Dis. 2017 Oct;23(7):828-839. doi: 10.1111/odi.12589. Epub 2016 Nov 24. Oral Dis. 2017. PMID: 27717092 Review.
-
Healthy Patients Are Not the Best Controls for Microbiome-Based Clinical Studies: Example of Sjögren's Syndrome in a Systematic Review.Front Immunol. 2021 Jul 29;12:699011. doi: 10.3389/fimmu.2021.699011. eCollection 2021. Front Immunol. 2021. PMID: 34394092 Free PMC article.
-
Understanding the roles of the microbiome in autoimmune rheumatic diseases.Rheumatol Immunol Res. 2023 Dec 19;4(4):177-187. doi: 10.2478/rir-2023-0027. eCollection 2023 Dec. Rheumatol Immunol Res. 2023. PMID: 38125641 Free PMC article.
-
[Psychiatric manifestations of lupus erythematosus systemic and Sjogren's syndrome].Encephale. 2001 Nov-Dec;27(6):588-99. Encephale. 2001. PMID: 11865567 French.
-
Sjögren's syndrome and systemic lupus erythematosus: links and risks.Open Access Rheumatol. 2019 Jan 29;11:33-45. doi: 10.2147/OARRR.S167783. eCollection 2019. Open Access Rheumatol. 2019. PMID: 30774485 Free PMC article. Review.
Cited by
-
Oral Microbiome: A Review of Its Impact on Oral and Systemic Health.Microorganisms. 2024 Aug 29;12(9):1797. doi: 10.3390/microorganisms12091797. Microorganisms. 2024. PMID: 39338471 Free PMC article. Review.
-
Analysis of Human and Microbial Salivary Proteomes in Children Offers Insights on the Molecular Pathogenesis of Molar-Incisor Hypomineralization.Biomedicines. 2022 Aug 24;10(9):2061. doi: 10.3390/biomedicines10092061. Biomedicines. 2022. PMID: 36140166 Free PMC article.
-
Oral Microbiota Signatures in the Pathogenesis of Euthyroid Hashimoto's Thyroiditis.Biomedicines. 2023 Mar 26;11(4):1012. doi: 10.3390/biomedicines11041012. Biomedicines. 2023. PMID: 37189630 Free PMC article.
-
Impact of oral microbiota on pathophysiology of GVHD.Front Immunol. 2023 Mar 9;14:1132983. doi: 10.3389/fimmu.2023.1132983. eCollection 2023. Front Immunol. 2023. PMID: 36969182 Free PMC article. Review.
-
Is poor oral health a risk factor for idiopathic granulomatous mastitis?Biomol Biomed. 2024 Sep 6;24(5):1310-1318. doi: 10.17305/bb.2024.10324. Biomol Biomed. 2024. PMID: 38488700 Free PMC article.
References
-
- Aletaha D., Neogi T., Silman A. J., Funovits J., Felson D. T., Bingham C. O., et al. . (2010). 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 62, 2569–2581. 10.1002/art.27584 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

