miR-539-5p regulates Srebf1 transcription in the skeletal muscle of diabetic mice by _targeting DNA methyltransferase 3b
- PMID: 36090753
- PMCID: PMC9439965
- DOI: 10.1016/j.omtn.2022.08.013
miR-539-5p regulates Srebf1 transcription in the skeletal muscle of diabetic mice by _targeting DNA methyltransferase 3b
Abstract
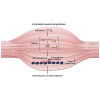
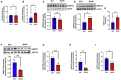
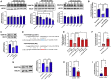
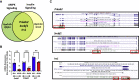
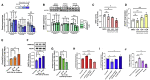
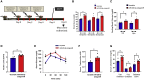
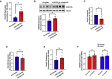
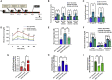
Aberrant DNA methylation is associated with diabetes, but the precise regulatory events that control the levels and activity of DNA methyltransferases (DNMTs) is not well understood. Here we show that miR-539-5p _targets Dnmt3b and regulates its cellular levels. miR-539-5p and Dnmt3b show inverse patterns of expression in skeletal muscle of diabetic mice. By binding to the 3' UTR of Dnmt3b, miR-539-5p downregulates its levels in C2C12 cells and in human primary skeletal muscle cells. miR-539-5p-Dnmt3b interaction regulates Srebf1 transcription by altering methylation at CpG islands within Srebf1 in C2C12 cells. Dnmt3b inhibition alone was sufficient to upregulate Srebf1 transcription. In vivo antagonism of miR-539-5p in normal mice induced hyperglycemia and hyperinsulinemia and impaired oral glucose tolerance. These mice had elevated Dnmt3b and decreased Srebf1 levels in skeletal muscle. db/db mice injected with miR-539-5p mimics showed improved circulatory glucose and cholesterol levels. Oral glucose tolerance improved together with normalization of Dnmt3b and Srebf1 levels in skeletal muscle. Our results support a critical role of miR-539-5p and Dnmt3b in aberrant skeletal muscle metabolism during diabetes by regulating Srebf1 transcription; modulating the miR-539-5p-Dnmt3b axis might have therapeutic potential for addressing altered skeletal muscle physiology during insulin resistance and type 2 diabetes.
Keywords: DNA methylation; DNMT3b; Srebf1; diabetes; miR-539-5p; skeletal muscle.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
H19 inhibition increases HDAC6 and regulates IRS1 levels and insulin signaling in the skeletal muscle during diabetes.Mol Med. 2022 Jul 16;28(1):81. doi: 10.1186/s10020-022-00507-3. Mol Med. 2022. PMID: 35842608 Free PMC article.
-
miR-135a _targets IRS2 and regulates insulin signaling and glucose uptake in the diabetic gastrocnemius skeletal muscle.Biochim Biophys Acta. 2013 Aug;1832(8):1294-303. doi: 10.1016/j.bbadis.2013.03.021. Epub 2013 Apr 8. Biochim Biophys Acta. 2013. PMID: 23579070
-
MiR-542-5p Inhibits Hyperglycemia and Hyperlipoidemia by _targeting FOXO1 in the Liver.Yonsei Med J. 2020 Sep;61(9):780-788. doi: 10.3349/ymj.2020.61.9.780. Yonsei Med J. 2020. PMID: 32882762 Free PMC article.
-
Reduced expression of microRNA-432-5p by DNA methyltransferase 3B leads to development of colorectal cancer through upregulation of CCND2.Exp Cell Res. 2022 Jan 1;410(1):112936. doi: 10.1016/j.yexcr.2021.112936. Epub 2021 Nov 19. Exp Cell Res. 2022. PMID: 34801563
-
DNA methylation of miR-181a-5p mediated by DNMT3b drives renal interstitial fibrosis developed from acute kidney injury.Epigenomics. 2024;16(13):945-960. doi: 10.1080/17501911.2024.2370229. Epub 2024 Jul 18. Epigenomics. 2024. PMID: 39023272 Free PMC article.
Cited by
-
An Updated Review on the Significance of DNA and Protein Methyltransferases and De-methylases in Human Diseases: From Molecular Mechanism to Novel Therapeutic Approaches.Curr Med Chem. 2024;31(23):3550-3587. doi: 10.2174/0929867330666230607124803. Curr Med Chem. 2024. PMID: 37287285 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

