_targeting the Heterogeneous Genomic Landscape in Triple-Negative Breast Cancer through Inhibitors of the Transcriptional Machinery
- PMID: 36139513
- PMCID: PMC9496798
- DOI: 10.3390/cancers14184353
_targeting the Heterogeneous Genomic Landscape in Triple-Negative Breast Cancer through Inhibitors of the Transcriptional Machinery
Abstract
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer defined by lack of the estrogen, progesterone and human epidermal growth factor receptor 2. Although TNBC tumors contain a wide variety of oncogenic mutations and copy number alterations, the direct _targeting of these alterations has failed to substantially improve therapeutic efficacy. This efficacy is strongly limited by interpatient and intratumor heterogeneity, and thereby a lack in uniformity of _targetable drivers. Most of these genetic abnormalities eventually drive specific transcriptional programs, which may be a general underlying vulnerability. Currently, there are multiple selective inhibitors, which _target the transcriptional machinery through transcriptional cyclin-dependent kinases (CDKs) 7, 8, 9, 12 and 13 and bromodomain extra-terminal motif (BET) proteins, including BRD4. In this review, we discuss how inhibitors of the transcriptional machinery can effectively _target genetic abnormalities in TNBC, and how these abnormalities can influence sensitivity to these inhibitors. These inhibitors _target the genomic landscape in TNBC by specifically suppressing MYC-driven transcription, inducing further DNA damage, improving anti-cancer immunity, and preventing drug resistance against MAPK and PI3K-_targeted therapies. Because the transcriptional machinery enables transcription and propagation of multiple cancer drivers, it may be a promising _target for (combination) treatment, especially of heterogeneous malignancies, including TNBC.
Keywords: bromodomain and extra-terminal (BET) proteins; transcription-associated cyclin-dependent kinases (CDKs); transcriptional machinery; triple-negative breast cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
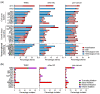

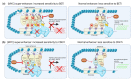
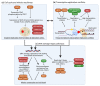

Similar articles
-
Kinase inhibitors for precision therapy of triple-negative breast cancer: Progress, challenges, and new perspectives on _targeting this heterogeneous disease.Cancer Lett. 2022 Oct 28;547:215775. doi: 10.1016/j.canlet.2022.215775. Epub 2022 Jun 3. Cancer Lett. 2022. PMID: 35667515 Review.
-
BRD4 modulates vulnerability of triple-negative breast cancer to _targeting of integrin-dependent signaling pathways.Cell Oncol (Dordr). 2020 Dec;43(6):1049-1066. doi: 10.1007/s13402-020-00537-1. Epub 2020 Oct 2. Cell Oncol (Dordr). 2020. PMID: 33006750 Free PMC article.
-
Bromodomain and Extraterminal Protein Inhibition Blocks Growth of Triple-negative Breast Cancers through the Suppression of Aurora Kinases.J Biol Chem. 2016 Nov 4;291(45):23756-23768. doi: 10.1074/jbc.M116.738666. Epub 2016 Sep 20. J Biol Chem. 2016. PMID: 27650498 Free PMC article.
-
Anti-tumor activity of BET inhibitors in androgen-receptor-expressing triple-negative breast cancer.Sci Rep. 2019 Sep 16;9(1):13305. doi: 10.1038/s41598-019-49366-9. Sci Rep. 2019. PMID: 31527644 Free PMC article.
-
A perspective on anti-EGFR therapies _targeting triple-negative breast cancer.Am J Cancer Res. 2016 Aug 1;6(8):1609-23. eCollection 2016. Am J Cancer Res. 2016. PMID: 27648353 Free PMC article. Review.
Cited by
-
Exploring the biological behavior and underlying mechanism of KITLG in triple-negative breast cancer.J Cancer. 2024 Jan 1;15(3):764-775. doi: 10.7150/jca.90051. eCollection 2024. J Cancer. 2024. PMID: 38213737 Free PMC article.
-
Systematic screening identifies ABCG2 as critical factor underlying synergy of kinase inhibitors with transcriptional CDK inhibitors.Breast Cancer Res. 2023 May 5;25(1):51. doi: 10.1186/s13058-023-01648-x. Breast Cancer Res. 2023. PMID: 37147730 Free PMC article.
-
Selective inhibition of CDK9 in triple negative breast cancer.Oncogene. 2024 Jan;43(3):202-215. doi: 10.1038/s41388-023-02892-3. Epub 2023 Nov 24. Oncogene. 2024. PMID: 38001268 Free PMC article.
References
-
- Bauer K.R., Brown M., Cress R.D., Parise C.A., Caggiano V. Descriptive Analysis of Estrogen Receptor (ER)-Negative, Progesterone Receptor (PR)-Negative, and HER2-Negative Invasive Breast Cancer, the so-Called Triple-Negative Phenotype: A Population-Based Study from the California Cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. - DOI - PubMed
-
- Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

