The contribution of functional HNF1A variants and polygenic susceptibility to risk of type 2 diabetes in ancestrally diverse populations
- PMID: 36216889
- PMCID: PMC9729131
- DOI: 10.1007/s00125-022-05806-2
The contribution of functional HNF1A variants and polygenic susceptibility to risk of type 2 diabetes in ancestrally diverse populations
Abstract
Aims/hypothesis: We examined the contribution of rare HNF1A variants to type 2 diabetes risk and age of diagnosis, and the extent to which their impact is affected by overall genetic susceptibility, across three ancestry groups.
Methods: Using exome sequencing data of 160,615 individuals of the UK Biobank and 18,797 individuals of the BioMe Biobank, we identified 746 carriers of rare functional HNF1A variants (minor allele frequency ≤1%), of which 507 carry variants in the functional domains. We calculated polygenic risk scores (PRSs) based on genome-wide association study summary statistics for type 2 diabetes, and examined the association of HNF1A variants and PRS with risk of type 2 diabetes and age of diagnosis. We also tested whether the PRS affects the association between HNF1A variants and type 2 diabetes risk by including an interaction term.
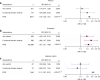
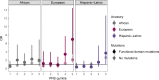
Results: Rare HNF1A variants that are predicted to impair protein function are associated with increased risk of type 2 diabetes in individuals of European ancestry (OR 1.46, p=0.049), particularly when the variants are located in the functional domains (OR 1.89, p=0.002). No association was observed for individuals of African ancestry (OR 1.10, p=0.60) or Hispanic-Latino ancestry (OR 1.00, p=1.00). Rare functional HNF1A variants were associated with an earlier age at diagnosis in the Hispanic-Latino population (β=-5.0 years, p=0.03), and this association was marginally more pronounced for variants in the functional domains (β=-5.59 years, p=0.03). No associations were observed for other ancestries (African ancestry β=-2.7 years, p=0.13; European ancestry β=-3.5 years, p=0.20). A higher PRS was associated with increased odds of type 2 diabetes in all ancestries (OR 1.61-2.11, p<10-5) and an earlier age at diagnosis in individuals of African ancestry (β=-1.4 years, p=3.7 × 10-6) and Hispanic-Latino ancestry (β=-2.4 years, p<2 × 10-16). Furthermore, a higher PRS exacerbated the effect of the functional HNF1A variants on type 2 diabetes in the European ancestry population (pinteraction=0.037).
Conclusions/interpretation: We show that rare functional HNF1A variants, in particular those located in the functional domains, increase the risk of type 2 diabetes, at least among individuals of European ancestry. Their effect is even more pronounced in individuals with a high polygenic susceptibility. Our analyses highlight the importance of the location of functional variants within a gene and an individual's overall polygenic susceptibility, and emphasise the need for more genetic data in non-European populations.
Keywords: Age at diagnosis; Functional domain; HNF1A; Interaction effects; Polygenic risk scores; Risk stratification; Type 2 diabetes.
© 2022. The Author(s).
Figures


Similar articles
-
Ancestry effects on type 2 diabetes genetic risk inference in Hispanic/Latino populations.BMC Med Genet. 2020 Jun 25;21(Suppl 2):132. doi: 10.1186/s12881-020-01068-0. BMC Med Genet. 2020. PMID: 32580712 Free PMC article.
-
Polygenic risk variants for type 2 diabetes susceptibility modify age at diagnosis in monogenic HNF1A diabetes.Diabetes. 2010 Jan;59(1):266-71. doi: 10.2337/db09-0555. Epub 2009 Sep 30. Diabetes. 2010. PMID: 19794065 Free PMC article.
-
Genome-wide polygenic risk score for retinopathy of type 2 diabetes.Hum Mol Genet. 2021 May 29;30(10):952-960. doi: 10.1093/hmg/ddab067. Hum Mol Genet. 2021. PMID: 33704450 Free PMC article.
-
Use of Polygenic Risk Scores for Coronary Heart Disease in Ancestrally Diverse Populations.Curr Cardiol Rep. 2022 Sep;24(9):1169-1177. doi: 10.1007/s11886-022-01734-0. Epub 2022 Jul 7. Curr Cardiol Rep. 2022. PMID: 35796859 Free PMC article. Review.
-
Perspectives on genetic studies of type 2 diabetes from the genome-wide association studies era to precision medicine.J Diabetes Investig. 2024 Apr;15(4):410-422. doi: 10.1111/jdi.14149. Epub 2024 Jan 23. J Diabetes Investig. 2024. PMID: 38259175 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- 200837/WT_/Wellcome Trust/United Kingdom
- F30 DK130576/DK/NIDDK NIH HHS/United States
- P30 DK020541/DK/NIDDK NIH HHS/United States
- R01 DK107786/DK/NIDDK NIH HHS/United States
- U01 DK105535/DK/NIDDK NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 DK124097/DK/NIDDK NIH HHS/United States
- P30 DK116074/DK/NIDDK NIH HHS/United States
- UM1 DK126185/DK/NIDDK NIH HHS/United States
- 200837/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 DK110113/DK/NIDDK NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

